Keywords
Cervical cancer, TLR9, Polymorphism, Genotypic frequency, Susceptibility
Cervical cancer, TLR9, Polymorphism, Genotypic frequency, Susceptibility
This version has been modified based on the reservations given by Dr. Gopeshwar Narayan and Prof. B. C. Das. The previous version of this article did not include the power calculation. In this version, we have calculated the statistical power based on the global minor allele frequency of the polymorphism under study and have drawn appropriate conclusions. The reason for the selection of TLR9 gene for this study has been mentioned in the Introduction as well as in the Discussion.
See the authors' detailed response to the review by Gopeshwar Narayan
See the authors' detailed response to the review by Bhudev C. Das
Cervical cancer is the fourth-most common cancer among women globally and second leading cause of cancer-related deaths in Indian women1. Although persistent infection of high-risk human papillomavirus (hrHPV) is considered as the chief causative agent of cervical cancer, variations in host genetic make-up does influence the risk of acquiring HPV infection, and susceptibility to cervical carcinogenesis2–4. In this context, variations in Toll-like receptor (TLR) genes, that play a crucial role in activating immune response by identifying pathogen-associated molecular patterns (PAMPs), have drawn significant attention, as single nucleotide polymorphisms (SNPs) in TLR genes have been shown to alter susceptibility to many infections and human diseases including cancer5–8.
Ten functional TLR genes are known in humans, the products of which recognizes specific PAMPs9. TLR1, TLR2, TLR4, TLR5 and TLR6 which are located on the cell surface of immune cells are known to recognize triacyl lipopeptides, peptidoglycans, lipopolysaccharides, flagellin and diacyl lipopeptides respectively that belong to bacterial cell wall or virus particles10,11. On the other hand TLR3, TLR7, TLR8 and TLR9 are located in the endosomal compartments, wherein TLR3 recognizes single as well as double stranded viral RNA, TLR7 recognizes ssRNA from viruses while the TLR9 gene product recognizes bacterial and viral DNA motifs including HPV16 CpG motifs5,10–12. Frequently analysed TLR9 SNPs G2848A and −1486 T/C have been suggested to alter cervical cancer susceptibility13–16, but no report is available elucidating the role of TLR9 Pro99Leu polymorphism in cancer. Although TLR9 Pro99Leu is a rare population SNP with a global minor allele frequency (MAF) of 0.0002 as reported in the single nucleotide polymorphism database (dbSNP), in-vitro analysis has revealed its significant role in DNA ligand hyporesponsiveness17. Considering the fact that cervical cancer is largely caused by hrHPV infection and TLR9 has the ability to respond to viral DNA, while other TLRs do not, the present study was designed to elucidate the association of the TLR9 Pro99Leu polymorphism with cervical cancer.
Biopsies from 110 cervical cancer patients and cervical smears from 141 healthy volunteers were collected from Shree Krishna Hospital, Anand; Sir Sayajirao General Hospital, Vadodara; and GMERS Hospital, Ahmedabad, India. The samples were collected from 2012 to 2017. The cancer biopsies and healthy cervical smears were histopathologically and cytologically confirmed. The clinical staging of cervical cancer samples was done as per The International Federation of Gynecology and Obstetrics (FIGO) guidelines.
DNA was isolated from cervical cancer biopsies and cervical smears by standard phenol-chloroform extraction method18. In the case of a low number of cervical cells, a spin-column based DNA isolation kit (Macherey-Nagel, Germany; Cat# 740952.50) was utilized as per manufacturer’s instructions. The quality and quantity of DNA was determined using ethidium bromide-stained 1% agarose gel on GelDoc system (BioRad, USA) as well as a NanoDrop 2000 (Thermofisher, USA). The TLR9 Pro99Leu polymorphism was detected using polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP) method as described by Kubarenko et al.17 Briefly, a 25µl PCR mix contained 0.1µM each of forward and reverse primer (Imperial Life Sciences, India), 0.1mM dNTP mix (Invitrogen, USA; Cat# 18427088), 2.5mM MgCl2 (Vivantis, USA; Cat# RB0204), 1 unit Taq DNA polymerase (Kapabiosystems, USA; Cat# KK1015) and 100 to 150ng genomic DNA. The PCR was run on an MJ Mini thermal cycler (BioRad, USA).
Upon confirmation of 337 bp PCR product on 2% ethidium bromide-stained agarose gel, 10µl PCR product was digested with BslI restriction enzyme (New England Biolabs, USA; Cat# R0555S) at 55°C overnight, separated on 12% polyacrylamide gel and analysed on a GelDoc system (BioRad, USA) for genotype identification. The details of PCR conditions and parameters for genotype consideration are mentioned in Table 1 and Table 2 respectively. To confirm the PCR-RFLP results, we performed Sanger sequencing of five randomly selected cervical cancer as well as control samples. All the sequencing reactions were performed on 3730xl DNA Analyzer (Applied Biosystems, USA) using BigDye™ Terminator v3.1 kit (Applied Biosystems, USA; Cat# 4337454) as per manufacturer’s instructions. The 10µl sequencing reaction was comprised of 7.0µl BigDye™ Terminator v3.1 Ready Reaction Mix, 10pmol forward primer and 50ng PCR product. The sequencing results were analyzed on Sequencing Analysis Software version 5.3.1 (Applied Biosystems, USA).
| Primer Sequence (5′ – 3′) | Thermal Profile | PCR Product | Visualized on |
|---|---|---|---|
| FP: GGATGTTGGTATGGCTGAGG RP: AACTGCAACTGGCTGTTCCT | (95°C – 5′) 1 (95°C – 45″, 56°C – 1′, 72°C – 30″) 35 (72°C – 10′) 1 | 337 bp | 2% Agarose Gel |
Statistical analysis was performed on GraphPad Prism version 5.00 for Windows (GraphPad Software, USA). Age of patients and controls were compared using two-sided Student's t-test. Due to the presence of single genotype across all the samples no additional statistical association was performed. The power of the study was calculated using Online Sample Size Estimator.
The average age of cervical cancer patients (52.43±11.78 years) and controls (51.8±11.35 years) was comparable without any statistically significant difference (p=0.668). Histopathologic analysis revealed all the cervical cancer cases to be of squamous cell carcinoma type. According to FIGO analysis, 9 (8.2%), 39 (35.5%), 55 (50%) and 7 (6.3%) patients belonged to Stage I, II, III and IV respectively. In the absence of polymorphic allele in the present population we calculated the power of the study based on the global minor allele frequency (0.0002) which was found to be 3.6%.
PCR amplification revealed the presence of a single intact band of 337 bp (Figure 1; Dataset 119). A single genotype CC (Pro/Pro) was detected across all the sample types (Table 3; Dataset 220) which was evident by the presence of 166 bp, 136 bp and 35 bp DNA bands after RFLP assay (Figure 2; Dataset 321). Sanger sequencing of the randomly selected PCR products corroborated with RFLP results (Figure 3; Dataset 422).
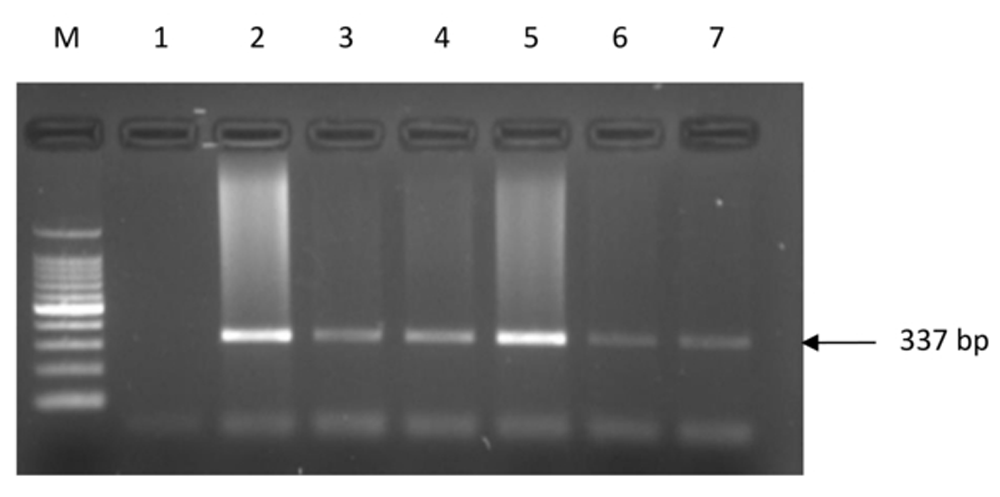
Lane M is 100 bp molecular marker (Takara, Japan; Cat# RR820A), Lane 1 is negative control and Lanes 2–7 are tumor DNA showing PCR products of 337 bp. (Abbreviations: PCR, Polymerase Chain Reaction; TLR9, Toll-like receptor 9; bp, base pair).
| Genotype | Cervical Cancer n (%) | Controls n (%) |
|---|---|---|
| CC | 110 (100.0) | 141 (100.0) |
| CT | 0 (0.0) | 0 (0.0) |
| TT | 0 (0.0) | 0 (0.0) |
| Allele | ||
| C | 220 (100.0) | 282 (100.0) |
| T | 0 (0.0) | 0 (0.0) |
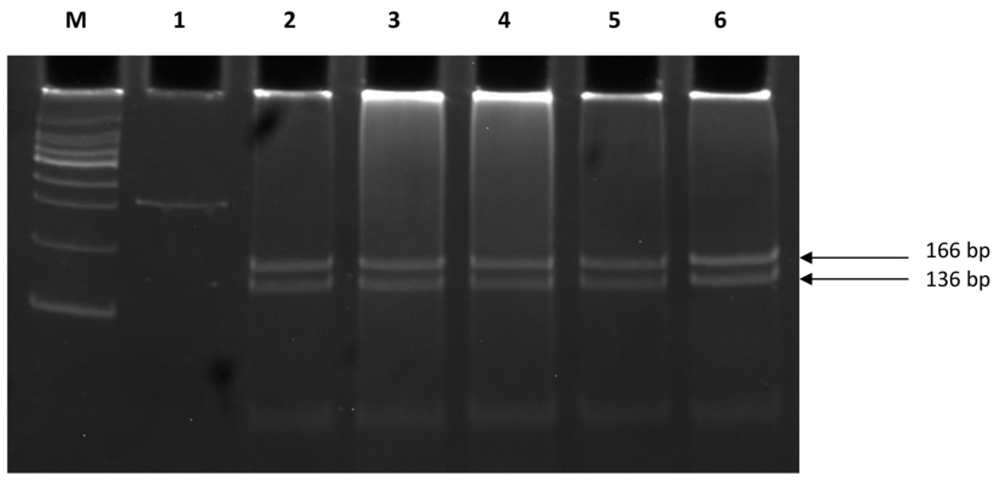
Lane M is 100 bp molecular marker, Lane 1 is undigested PCR product and Lanes 2 to 6 are showing digested PCR products of 166 bp and 136 bp (35 bp band is not visible) by BslI enzyme representing CC genotype. (Abbreviations: PAGE, Polyacrylamide Gel Electrophoresis; RFLP, Restriction Fragment Length Polymorphism).

Sanger sequence electropherogram of (A) a healthy individual and (B) patient showing single peak (highlighted) of C allele of TLR9 C296T/ Pro99Leu SNP representing CC genotype. (Abbreviations: SNP, Single Nucleotide Polymorphism).
Although hrHPV infection is the primary etiological agent of cervical carcinogenesis, the role of host genetic factors, especially those associated with body immunity such as TLRs, cannot be ignored. TLR9 which recognizes CpG DNA motifs from bacteria and viruses has also been reported to recognize HPV16 CpG DNA12. Moreover, variations in the TLR9 gene has found associations with various diseases including cancer. TLR9 SNPs −1486 T/C and G2848A have been found to be contradictorily associated with cervical cancer risk. In Polish and Mexican populations both TLR9 −1486 T/C and G2848A polymorphisms were suggested to be risk factors for cervical carcinogenesis13,15. In two independent studies on Chinese population, a positive association with TLR9 G2848A SNP was detected23,24 but no involvement of TLR9 −1486 T/C was found24, however, the other study suggested −1486 T/C was not a contributory factor to cervical carcinogenesis14. From India, a single report on North Indian patients revealed a marginal role of TLR9 G2848A polymorphism with cervical cancer risk16.
To date, no report is available on the rare TLR9 Pro99Leu polymorphism in cancer, which has been shown to be associated with DNA ligand hyporesponsiveness in HeLa cell lines17. Considering the fact that cervical cancer is majorly caused by hrHPV infection and the TLR9 Pro99Leu polymorphism is associated with DNA ligand hyporesponsiveness, the present study investigated, for the first time, the role of the TLR9 Pro99Leu polymorphism in cervical cancer susceptibility. This is also the first report to study this polymorphism in any of the cancer types globally. Our results revealed the presence of a single genotype CC (Pro/Pro) among cases and controls, demonstrating no significance of the Pro99Leu polymorphism to cervical cancer susceptibility. Similarly, no association with high-risk HPV infection, that was detected in almost 70% of the patients, was found (details of HPV infection to be published elsewhere). A complete absence of Pro99Leu in our study population corroborates with the report of Lee and group (2006) where neither controls nor lung tuberculosis and sarcoidosis patients had the TLR9 Pro99Leu polymorphism25. Similarly, the Pro99Leu polymorphism was not detected among healthy Caucasians as well as pneumococcal disease, bacteraemia, and leprosy patients17. Moreover, according to dbSNP, the global MAF of this polymorphism is 0.0002, and our results, albeit on a smaller cohort, do solicit its rare polymorphism status. Therefore, owing to sample size constraint, it would be inappropriate to draw a direct conclusion of the effect of above-said polymorphism on the susceptibility to cervical cancer. As the power of study calculated based on the global MAF was very low (3.6%), and to achieve a power of study of 80%, approximately 40000 samples will be required to analyze. Finally, under present scenario, a direct role of this SNP in cancer, as well as other diseases, seems a remote possibility. Nonetheless, a comprehensive analysis of a larger cohort covering a varied ethnic population globally is suggested to comprehend its role in microbial infection and/or disease susceptibility including cancer.
The preliminary data obtained from the present study does not suggest a role for the TLR9 Pro99Leu polymorphism in cervical cancer susceptibility. However, analysis on a larger cohort worldwide may provide more insights into the frequency distribution of Pro99Leu polymorphism and reveal its influential role in various human diseases including cancer.
Dataset 1. Raw agarose gel images of PCR amplification of TLR9 gene segment for C296T/ Pro99Leu polymorphism from 50 samples consisting of 26 controls and 24 cervical cancer cases. Figure 1 is a representative picture of the same. 10.5256/f1000research.14840.d20340519
Dataset 2. Age, clinical stage and TLR9 genotype status among cervical cancer patients as well as age and TLR9 genotype status among controls. 10.5256/f1000research.14840.d20340620
Dataset 3. Raw polyacrylamide gel electrophoresis images of 27 controls and 24 cervical cancer PCR amplified products that underwent restriction fragment length polymorphism (RFLP) analysis. Figure 2 is a representative picture of the same. 10.5256/f1000research.14840.d20340721
Dataset 4. Nucleotide sequences spanning TLR9 gene segment for C296T single nucleotide polymorphism, obtained after performing Sanger sequencing on five samples each of cervical cancer and healthy controls. The sequencing results confirm the restriction fragment length polymorphism (RFLP) analysis that represents single genotype CC among all the study subjects. Figure 3 A and B are representative electropherograms of the TLR9 C296T CC genotype as evident by the presence of single peak of C allele. 10.5256/f1000research.14840.d20340822
The research was carried out following due approval from ethics committee of all the participating institutes. Subjects were verbally informed and explained about the study, and were provided with an information sheet. Written informed consent was obtained from the subjects who agreed to enrol in the present study. Personal information of all the study subjects was kept confidential.
The study was funded by Charotar University of Science and Technology (CHARUSAT).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authors thank Dr. Anjana Chauhan, Gynec Cancer Surgeon, Ex Associate Professor, Gujarat Cancer and Research Institute, Ahmedabad, India for stimulating discussions.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 30 Aug 18 |
read | read | read |
|
Version 1 17 May 18 |
read | read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)