Keywords
Psychrophilic fungi, phenotype microarray, metabolism, catabolism, gene function
This article is included in the Pathogens gateway.
Psychrophilic fungi, phenotype microarray, metabolism, catabolism, gene function
We modified/added a number of items in response to the reviewers' comments. The main changes are:
See the authors' detailed response to the review by Christopher T Cornelison
See the authors' detailed response to the review by Flavia Pinzari
See the authors' detailed response to the review by Christine Salomon
Pseudogymnoascus destructans causes white-nose syndrome (WNS), a disseminated disease afflicting hibernating bats in North America since 20061–3. WNS is linked to mass mortality and now afflicts bats over large geographic areas in the United States and Canada. P. destructans’ pathogenic mechanisms remain mysterious especially as no other human or animal fungal pathogen expresses virulence attributes at such low temperatures. Pseudogymnoascus pannorum, a closely related fungus, is widely distributed in the soil and substrates of caves and mines in North America3. P. pannorum grows both at psychrophilic and mesophilic temperature ranges and causes human and canine diseases rarely4. However, P. pannorum does not cause any disease in hibernating bats. These facts raise the exciting possibilities that P. destructans is more specialized for the pathogenic lifestyle on bats while P. pannorum successfully colonizes a broader range of substrates in nature.
Environmental studies on the psychrophilic and psychrotolerant fungi documented the versatility of Pseudogymnoascus (Geomyces) pannorum for the utilization of complex carbohydrates and keratin-enriched substrates, and tolerance to high salt5–7. Additional laboratory studies demonstrated extensive saprotrophic enzymatic activities that would allow resource capture by the non-pathogenic Pseudogymnoascus species vis-a-vis P. destructans8,9. P. destructans is known to secrete proteolytic, lipolytic, and keratinolytic exoenzymes, and possesses specialized catabolic activities that contribute to its growth and survival in the nutrient-poor caves and mines2,10.
Although their draft genomes are similar in size (~30 Mb), there are numerous repeats and far fewer proteins and enzymes in P. destructans (2,052 proteins) than in P. pannorum (2,734 proteins)11. In the present study, we report the results of extensive Biolog Phenotype Microarray metabolic profiling to confirm in silico gene predictions, and find clues for the different lifestyles of these psychrophilic fungi.
The metabolic analysis was conducted using P. destructans (M1379) and P. pannorum (M1372)11. The PM1-10 and PM21, 23–25 phenotype microarray plates were procured from Biolog, Hayward, CA. The fungal spores were harvested in sterile water from 3 - 5-week-old, heavily sporulating culture on potato dextrose agar (PDA) flasks at 15°C. In preliminary experiments, spore counts and viability were determined on agar plates using a hemocytometer and colony forming units (CFU). For the final tests, the spores were harvested, washed once in sterile water by centrifugation, and the suspension adjusted to an OD600 = 0.2 (transmittance = 62%). This suspension equated to between 550 and 950 spores per well via hemocytometer count, and 250–500 spores per well by CFU. In preliminary experiments, the two fungi grew at different growth rates and comparable growth was observed after day 7 for P. pannorum and day 10 for P. destructans (details not shown). Further incubation of the plates beyond the observation period did not change the observed growth pattern.
The PM plates were inoculated per Biolog protocol and incubated at 15°C12,13. The presence or absence of growth was measured by OD600 on day 10 for P. destructans, and day 7 for P. pannorum. Negative control wells were weakly growth positive for both P. destructans and P. pannorum. This observation was also reported for Biolog PM plates in another study13. Therefore, the corresponding negative control well reading from each experiment were averaged together and used to normalize the OD values averages for each test compound. For the heat map visualization, the negative control reading was assigned a score of 0.0 and the positive growth scored on a 0.0 – 1.0 scale. The phenotypic assay was repeated once. The limited dataset precluded any quantitative statistical analysis.
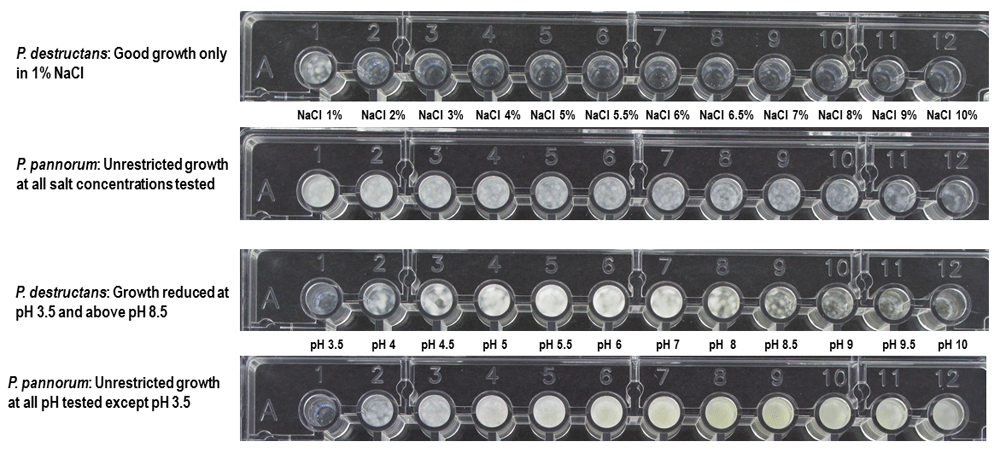
Nearly 1,047 different metabolic activities were analyzed for each test fungus (Datasets 1–414). P. pannorum metabolized far more carbon and nitrogen compounds; P. destructans exhibited prominent activity on phosphorous sources and nutrient supplements (Figure 1). P. pannorum utilized 78 of 190 carbon sources (41%), and 41 of 91 nitrogen sources (43%) tested. P. destructans used 23 carbon compounds (12%) and 23 nitrogen compounds (24%). P. destructans exhibited more robust growth on the phosphorous sources and nutrient supplements (83% and 15%, respectively) compared to P. pannorum (27% and 1%, respectively.). P. pannorum metabolized nearly all carbon intermediates in the major fungal metabolic cycles13 (Figure 2). P. destructans utilized only a few simple sugars in glycolysis with no activity on a range of carbon intermediates. P. pannorum used a wider variety of nitrogen sources including amino acids, amino bases, and alkanes while P. destructans had a preference for the simple N sources and dipeptides13 (Figure 3). Most phosphorous sources tested supported the growth of P. destructans while P. pannorum only grew on few phosphosugars and phosphorylated nucleosides (Figure 4). Both fungi did not utilize sulfur intermediates (Datasets 1–414). Fifteen of ninety-five nutrient supplements supported good growth of P. destructans while P. pannorum grew only on D-Pantothenic acid (Supplementary files). P. pannorum grew at very high salt concentrations and extreme acidic and basic pH ranges while P. destructans was sensitive to high salt (diminished growth ≥ 1% NaCl) and basic pH (diminished growth > pH 8.5) (Figure 5). P. pannorum showed extreme tolerance to 96 xenobiotics in PM21, PM23 - PM25 plates in contrast to severe sensitivity observed in P. destructans (details not shown).
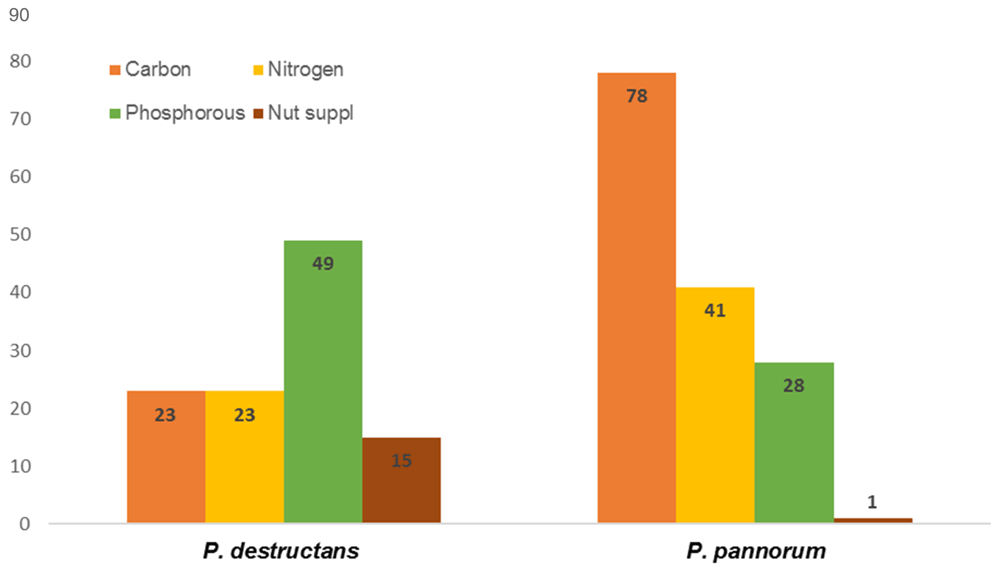

The details of test set-up and end point reading are described in the methods. For the heat map visualization, the negative control reading was assigned a score of 0.0 and positive growth scored on a 0.0 – 1.0 scale.
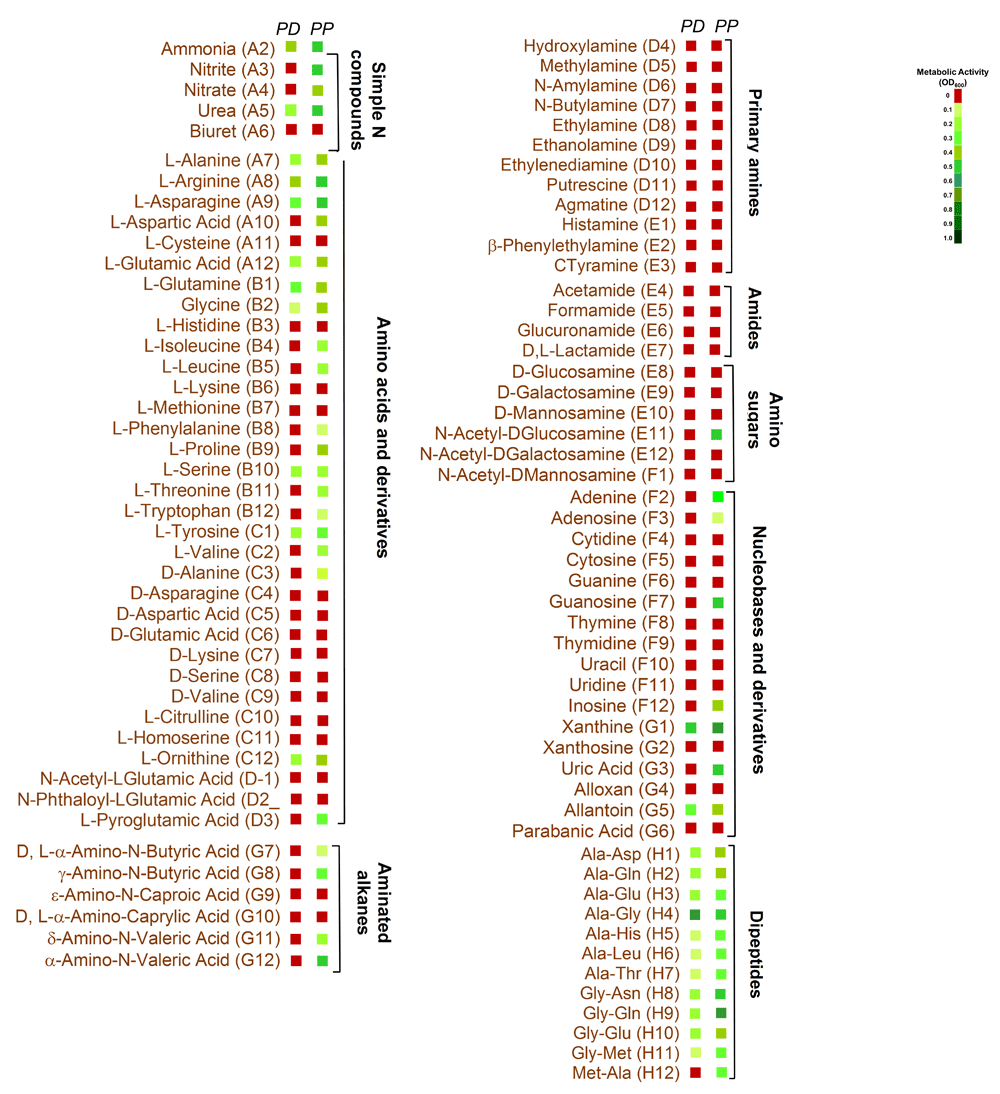
The details of test set-up and heat map are similar to Figure 2.
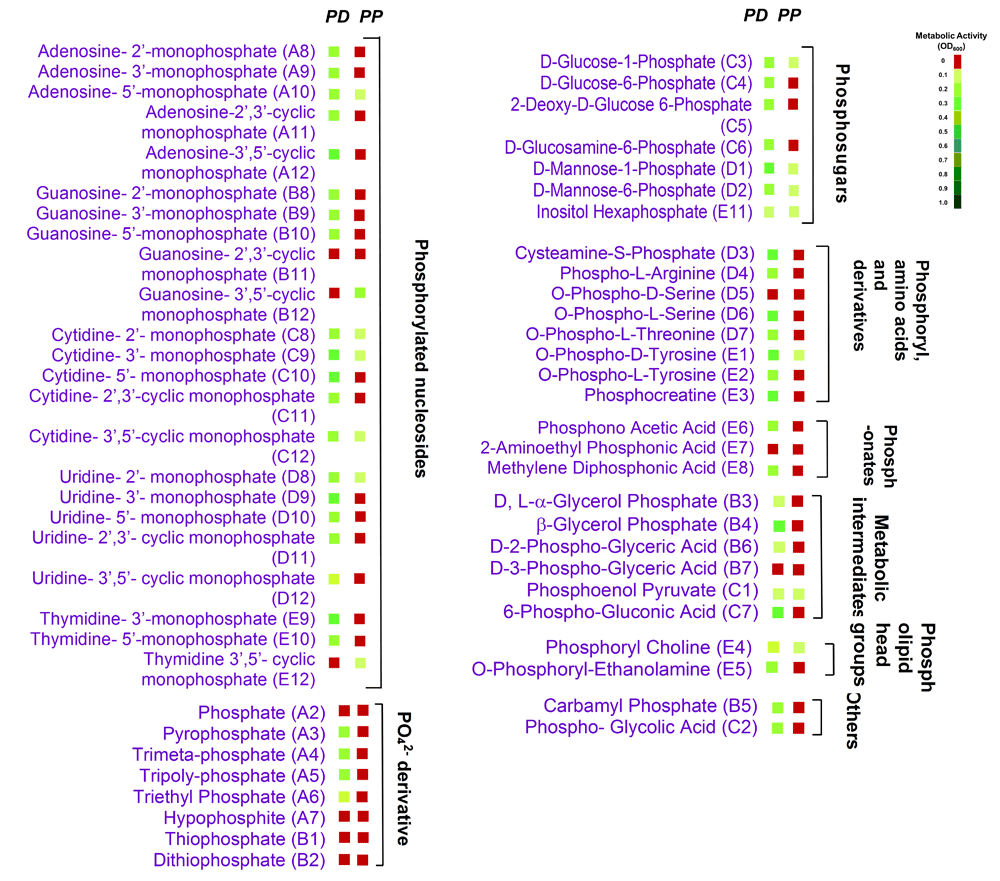
The details of test set-up and heat map are similar to Figure 2.
Metabolic profiles of P. destructans and P. pannorum validated in silico predictions about the notable differences in the number of protein-encoding genes in their genomes11. P. destructans contained enzymes and catabolic pathways that support fungal growth on a limited range of substrates of non-plant origin and showed high sensitivity to stress. P. pannorum was remarkably adapted for the nutrient poor environments of the caves and mines (‘extremophile’) with oligotrophic metabolism, osmotolerance, xerotolerance, and xenobiotic tolerance.
The findings in the present study confirm and expand on results from other reports on P. destructans’ adaptation and persistence in the North American caves and mines in the face of possible competitive interactions with the native fungal species8–10. Both Raudabaugh and Miller (2013) and Reynolds and Barton (2014) used a variety of biochemical tests to probe the metabolic activities in a collection of Pseudogymnoascus species isolates9,10. The authors of the former study surmised the suitability of P. destructans as a saprobe in the affected caves and mines in limited biotic competition (‘resource island’)10. Reynolds and Barton (2014) found a reduced saprotrophic ability in P. destructans isolates vis-à-vis P. pannorum and other Pseduogymnoascus species, which suggested ‘co-evolution with the host’9. Wilson et al. (2017) performed a variety of tests including Biolog FF Microplate with 95 different substrates, and found limited saprotrophic ability in P. destructans in comparison to other Pseudogymnoascus species8.
Further Phenotype Microarray profiling of P. destructans and P. pannorum would be crucial to fill-in current gaps in their genome sequences, define gene functions, and elucidate pathophysiological attributes11,15,16.
The limitations of the current study include the use of single strains of two fungal species, and single end points instead of growth curves, which allow curve analysis for more accurate data interpretation as highlighted by other investigators.
We and others hope to accomplish these milestones with the recent availability of a high-quality P. destructans genome and data pipelines to automate Biolog analysis15,17–20.
Datasets 1–4: Excel sheets with OD600 values for all Biolog plates tested in this study. DOI, 10.5256/f1000research.15067.d20467914
This study was supported by the National Science Foundation (Award Number 1203528).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Microbial ecology, microbial natural products chemistry, fungal and bacterial infectious disease, biological control
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Canfora L, Abu-Samra N, Tartanus M, Łabanowska BH, et al.: Co-inoculum of Beauveria brongniartii and B. bassiana shows in vitro different metabolic behaviour in comparison to single inoculums.Sci Rep. 2017; 7 (1): 13102 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Emerging fungal pathogens, microbial control, applied microbiology.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Microbial ecology, microbial natural products chemistry, fungal and bacterial infectious disease, biological control
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 17 Jul 18 |
read | read | |
|
Version 1 25 May 18 |
read | read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)