Keywords
Network Biology, Shortest Paths, Pathway Reconstruction, CyREST API
This article is included in the Cytoscape gateway.
This article is included in the Bioinformatics gateway.
Network Biology, Shortest Paths, Pathway Reconstruction, CyREST API
PathLinker is an algorithm that automates the reconstruction of any human signaling pathway by connecting the receptors and transcription factors (TFs) in that pathway through a physical and regulatory interaction network1. In a comprehensive quantitative evaluation on NetPath pathways, PathLinker achieved higher reconstruction accuracy than several other state-of-the-art algorithms. In addition, PathLinker’s novel prediction that the cystic fibrosis transmembrane conductance regulator (CFTR), an ion-channel receptor, is involved in the Wnt pathway was experimentally validated1. In general, PathLinker can be used to connect any set of sources to any set of targets in a given network.
The PathLinker app for Cytoscape is an implementation of this algorithm. The PathLinker app2 was first released in the middle of December 2015. While it is possible to use the PathLinker app in conjunction with other Cytoscape apps, it can be cumbersome to create such workflows using the Cytoscape user interface. It is also challenging to reproduce the results of these workflows.
Here we present an CyREST-based API that allows users to incorporate PathLinker algorithms into their own software pipelines. The PathLinker application programming interface (API) will facilitate automated analysis of complex networks in reproducible workflows, including in conjunction with other CyREST enabled Cytoscape apps3.
The PathLinker API allows external software (written in languages such as Python and R) and tools (e.g., Jupyter Notebook) to access PathLinker functions via the REST protocol. The API follows the OSGi design pattern. The PathLinker API exposes two functions via JAX-RS annotations that allows them to be discovered by CyREST3 and be made available to external callers via REST. We have documented these functions using Swagger annotations to meet the Cytoscape Automation documentation standards. The Swagger annotation allows the PathLinker API functions to be accessed via the Swagger user interface along with other API functions exposed by CyREST.
We substantially updated and refactored the PathLinker codebase to follow the principles of the OSGi modular design and to remove redundant code. Specifically, we refactored the code for generating the k shortest paths, network visualization, and functions related to the user interface (e.g., generating information in the “Result Panel”) into distinct Task classes that can be managed by the Task Manager in the Cytoscape API. The current design enables us to use the same codebase for running PathLinker through the Cytoscape user-interface as well as through the REST API. This modular design will facilitate easy expansion of the app in the future, e.g., by implementing additional subnetwork-finding algorithms.
The PathLinker API is accessible directly through the Swagger user interface within Cytoscape or by using any REST-enabled client. Note that users must have installed v1.4 of the PathLinker app and Cytoscape v3.6.0 or higher. Moreover, an instance of Cytoscape must be running on the user’s computer. In the “Use Cases” section, we describe a sample workflow using py2cytoscape and provide an example Jupyter Notebook.
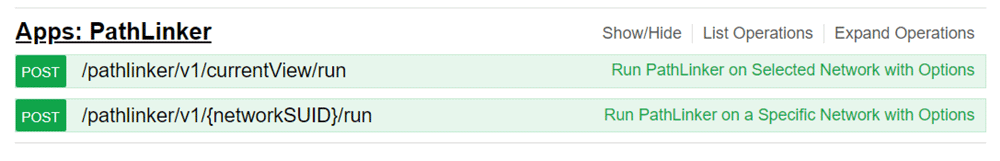
As shown in Figure 2, the PathLinker API provides two POST functions, “/pathlinker/v1/currentView/run” and “/pathlinker/v1/networkSUID/run”, which run PathLinker on the currently selected network and on the given networkSUID respectively. Both functions send user-selected source nodes and target nodes and a set of parameters to the PathLinker Cytoscape app.
The app computes the k shortest simple (loopless) paths that connect any source to any target in the network specified in the POST function, generates a subnetwork that contains these paths and a view of this subnetwork, creates a table in the Result Panel that contains these paths (See Figure 3b), and adds a “path rank” column to the Edge Table that contains the rank of the first path in which each edge appears. The POST functions return the computed paths, the SUIDs of the subnetwork and subnetwork view, and the name of the “path rank” column created by the app.
We summarize the parameters of the API functions and their outputs below.
API parameters
The API functions have the same set of parameters as the user can set in the user interface of the PathLinker app (see Figure 1a and Figure 1b). The user should provide these parameters in JSON format, whether they use the Swagger user interface or invoke the PathLinker API in code or via external tools. Table 1 contains an overview of the parameters, their types and a brief description. For a detailed description of the parameters, please see the Swagger documentation or documentation of the PathLinker app.
The last column records whether an equivalent option is present in the user interface of the PathLinker app. T/F - True/False.
API Output
The PathLinker API functions returns a response in JSON format with the following fields:
subnetworkSUID: The SUID of the subnetwork created in Cytoscape.
subnetworkViewSUID: The SUID of the subnetwork view created in Cytoscape.
pathRankColumnName: The name of the edge column created in the subnetwork’s edge table containing the rank of the first path in which each edge appears. The user can programmatically access this table using the CyREST API.
paths: The list of paths generated by the algorithm sorted by the path rank. This list contains the same information as the result table in the “Result Panel” created by the PathLinker app. Each entry in the list contains the following fields that describe a path:
rank: The rank of the path. For example, “100” for the 100th shortest path.
score: The total weight of the edges in the path.
nodeList: The list of nodes in the path. Each element in this list appears in the “name” column in the node table of the network provided as input to PathLinker. If multiple paths have the same score, then we order them lexicographically by their node lists.
If the user sets skipSubnetworkGeneration to true in the input to the API functions, then the PathLinker app does not generate the subnetwork and its view. Therefore, the API output will not contain the first three attributes, i.e., subnetworkSUID, subnetworkViewSUID, and pathRankColumnName.
The functions in the PathLinker API implement careful checks of the input. The function return the following error codes to provide meaningful feedback to the user:
400: Invalid user input. The JSON response lists all faulty input along with a reason why the input is correct, allowing the user to correct all the errors at the same time.
404: Current network or network with the given SUID does not exist.
422: No paths found. This error message indicates that PathLinker could not find any path connecting the source(s) to the target(s) in the given network. This error can occur if every source is disconnected from every target.
We provide two Jupyter Notebooks that illustrate the PathLinker API. The first notebook implements a use case on a simple network with 11 nodes and 13 edges, shown in Figure 3a. The notebook shows how to use Python and the py2cytoscape library to 1) load this network into Cytoscape, 2) call the PathLinker API with a set of parameters (Figure 1), 3) view the computed paths and subnetwork, and 4) save the paths and/or subnetwork image to a file. The API function call in the notebook asks PathLinker to find the two shortest paths connecting “a” to “e” or to “h” while treating the network as undirected. It also asks PathLinker to return more than two paths if their scores are tied with the second path.
Even though we set k = 2, PathLinker returned three paths; Figure 3b shows the subnetwork containing these paths. PathLinker did so because we used the “Include tied paths” option, and the second and third path both had a score of three (they contain three edges each). In this example, we used the “Treat network as undirected” option because even though the edges in the network were intended to be undirected, py2cytoscape treats networks imported from the Python NetworkX package as directed.
The second notebook implements a more complex example that we presented in the paper describing the PathLinker app2. Here, we used PathLinker to compute and analyze a network of interactions connecting proteins that are perturbed by the drug Lovastatin. The PathLinker API functions now allow us to automate this use case enabling fast reproduction of the results, as shown in the Jupyter Notebook.
The subnetwork returned by the PathLinker API is another network in the current Cytoscape session. Hence, this network is amenable for analysis by other CyREST APIs and/or Cytoscape apps. Some examples include calling the Diffusion app’s API on the subnetwork, or modifying the subnetwork using standard CyREST API calls such as adding visual styles to the subnetwork or calling different layout algorithms.
In the near future, we plan to provide users more freedom in determining which column in the Node Table should act as the input for source and target fields. We will also implement additional sub-network generation algorithms. We will implement these new features in parallel in the PathLinker app and in the REST API.
We hope that these additions will increase making PathLinker a method of choice in the network biology community.
All data underlying the results are available as part of the article and no additional source data are required.
Software available from:http://apps.cytoscape.org/apps/pathlinker
The Cytoscape app source code and sample Jupyter Notebooks are available at https://github.com/Murali-group/PathLinker-Cytoscape
Archived source code as at time of publication: http://dx.doi.org/10.5281/zenodo.12523084
License: GNU General Public License version 3
National Science Foundation grant [CCF-1617678] supported this research. The research is also partially based upon work supported by the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA), via the Army Research Office (ARO) under cooperative Agreement Number [W911NF-17-2-0105]. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the ODNI, IARPA, ARO, or the U.S. Government. The U.S. Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright annotation thereon.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Systems biology, bioinformatics
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 12 Jun 18 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)