Keywords
Open PHACTS, drug discovery, semantic, bioinformatics, WikiPathways, pathway database, API
Open PHACTS, drug discovery, semantic, bioinformatics, WikiPathways, pathway database, API
See the authors' detailed response to the review by Augustin Luna
See the authors' detailed response to the review by Yi-An Chen
Targeting proteins to ideally restore normal biological processes is a common starting point in drug discovery1. The Open PHACTS Discovery Platform (OPDP) was designed to help identify protein targets and information about their associations with each other2–4. The OPDP supports target identification and validation by including target-target interactions from WikiPathways5–7. Of these interaction networks, proteins sharing a downstream path allows investigation of alternative drug target combinations. Even the knowledge of which biological pathways participate in disease-related processes provides insight in the pathway topology between the targets. The importance and need of providing access to interaction information for real-world research questions was outlined in a recent Open PHACTS paper8.
The Open PHACTS project was born out of the desire to integrate pharmacological data from multiple precompetitive sources to efficiently address scientific questions that cannot be answered with single data sources8. It integrates data using linked data approaches3 from chemical and biological sources such as ChEBI, ChEMBL, UniProt, and WikiPathways6. However, the OPDP did not previously include calls to access specific up- and downstream interaction effects. This information is needed for questions related to drug repositioning and repurposing. Up- or downstream targets may be interesting alternatives with similar therapeutic effect to targets, for which it is particularly hard to develop a drug agent. Thus, finding a target that has already been drugged or is more drug tractable will be advantageous. Here we describe how to identify alternative targets in the same cellular pathway using OPDP against the WikiPathways data.
The WikiPathways Resource Description Framework data (WPRDF) is released as part of the monthly releases5. The native format for WikiPathways is Graphical Pathway Markup Language (GPML) based on the eXtensible Markup Language (XML) standard. The RDF export is transformed from the original GPML. In the RDF representation we use two distinct controlled vocabularies, to distinguish between the graphical notation of a pathway and the biological meanings expressed in the pathway. This is done to allow integration with other pathway repositories which use other graphical notations or none. The WikiPathways RDF also includes details about directed and undirected interactions. Directed biochemical interactions capture the source and target which are depicted as an arrow in simple pathway drawings. WikiPathways adds biological meaning to interactions with Molecular Interaction Map (MIM) interaction types, like inhibitions, enzyme catalyzed reactions, and stimulations9, as well as Systems Biology Graphical Notation (SBGN) interactions10. Reactome pathways in WikiPathways use SBGN interactions11,12. However, because MIM and SBGN use different drawing styles, we normalize their inhibition types into a common inhibition type, defined by the WikiPathways ontology (https://vocabularies.wikipathways.org/wp).
The WikiPathways basic drawing tools also contain generic arrows and T-bar annotations that give the user the ability to create basic diagrams without the semantic meaning of MIM or SBGN notations. The interactions connecting these nodes are captured, but the only explicit information is that it is a directed interaction from a source to a target. To handle more complicated enzyme reaction drawings, where there is not a single line that directly connects targets in a cascade of enzymatic reactions, a query was developed that recognizes these types of reactions. However, this is not implemented in the current Open PHACTS Application Programming Interface (API).
Version 2.1 of the OPDP API contains three new calls for interactions and their pathways. The first call, /pathway/getInteractions, returns all interactions involved in a pathway. To use this feature, the user specifies a pathway URI and OPDP returns its interactions including information about direction and the connected entities. The direction information is relayed as a starting node having a wp:source annotation, while the end of the interaction has the wp:target annotation. In its simplest form, this means that if gene product A is interacting with a gene product B, then we have wp:source for product A and wp:target for product B. However, the presented new methods also support interactions with multiple sources and targets for more complex interactions that are more accurately represented this way.
The second added call, /pathways/interactions/byEntity, returns the direction of the interactions involving this entity. An entity is specified by a URI and can be a metabolite, protein, gene product, or RNA. API options allow the user to select only upstream or only downstream interactions. If a direction is not specified in the call, all the adjacent interactions will be retrieved regardless of their direction. The results also specify the interaction type (e.g. inhibition, stimulation, conversion). Vocabularies.wikipathways.org also identifies catalysis and binding events as well as a more generic directedInteraction in the case where the type of the interaction is not identified. This ability to select the interaction direction is specifically what allows users to answer scientific questions around upstream and downstream effects, such as those defined by Open PHACTS. The third API call is /pathways/interactions/byEntity/count which is a helper function that returns the number of interactions for a target.
The OPDP API calls are backed by SPARQL searches against the loaded WikiPathways RDF. The query parameters that are required or optional are given in the documentation of Open PHACTS (https://dev.openphacts.org/docs/2.1). As in previous versions, the API uses HTTP GET to call methods and needs a (free) application ID and key (see https://dev.openphacts.org/signup)3.
To ensure multiple URI schemes can be used to identify genes, proteins, and metabolites, the Open PHACTS platform uses an Identifier Mapping Service (IMS)6. This ensures that people can use Ensembl, NCBI Gene, and others for genes, UniProt, Ensembl, etc. for proteins, and HMDB, ChEBI, CAS registry number, and PubChem for metabolites. Furthermore, it supports identifiers.org formatted URIs, further simplifying entering identifiers13.
We are demonstrating the platform with three example calls. All the API calls require use of an application ID and an application key. This key and ID can be acquired by creating a free Open PHACTS account. The first example is an application to the PI3K/AKT pathway for cell growth regulation which contain important targets for cancer treatment14. The AKT protein has a central role and usefully shows the API call’s ability to return connected elements with the /pathways/interactions/byEntity and the /pathway/getInteractions calls. The API calls can help aid drug discovery by taking a target, in this case AKT, and easily identify other connected proteins that could potentially be used as drug targets with a common downstream effect.
Figure 1 shows the web interface of the API call that returns the connectivity of the AKT2 target to both upstream or downstream proteins or gene products. This method allows the user to identify connections to other targets in the pathway. The results of that API call (Figure 2) show the AKT2 interaction with microRNA. A helper method (Figure 3): /pathways/interactions/byEntity/count is also included. It returns the number of all interactions in which an entity is participates. This helps the user get a sense of the prevalence of the queried entity with interactions in pathways found on WikiPathways. An example result for this query can be found in Supplementary Figure 1.
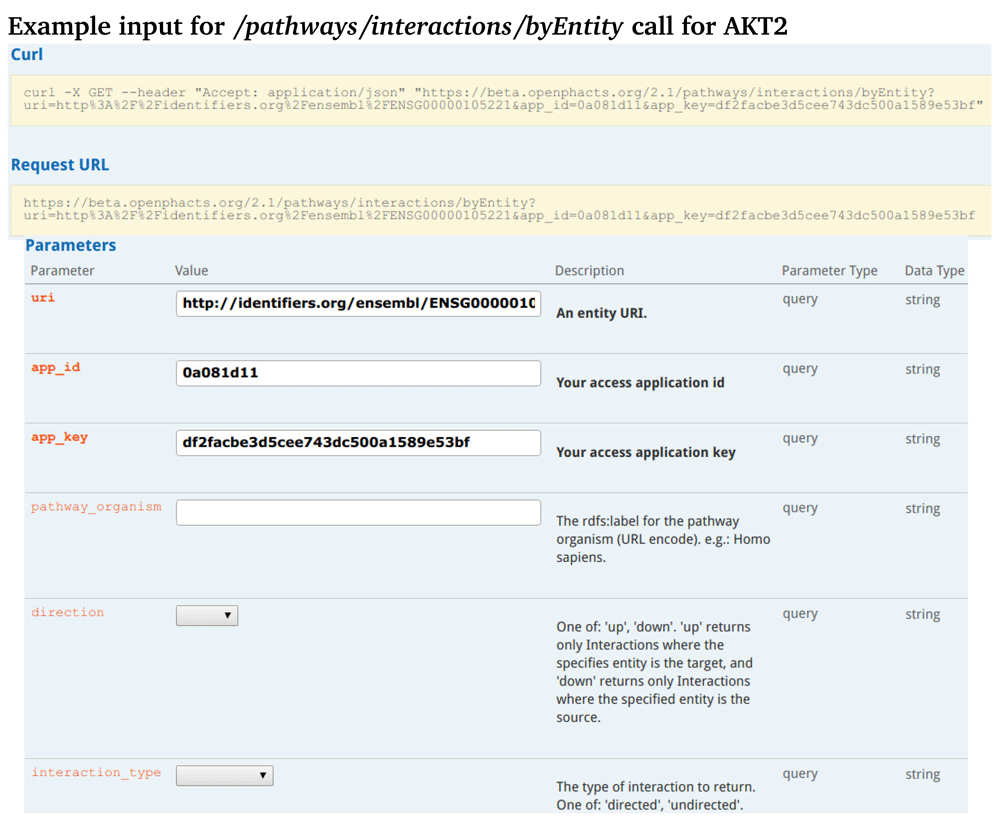
The GET portion tells the API to retrieve data with the associated call. It takes an entity URI, the Ensembl ID for AKT2, and returns a list interactions for AKT2. The obligatory parameters are shown in bold. Entity IDs that are acceptable for queries include Ensembl, Entrez Gene, and UniProt for genes, proteins, and RNAs. For metabolites the ID sources HMDB, ChEBI, and ChemSpider, for example, are acceptable entity IDs
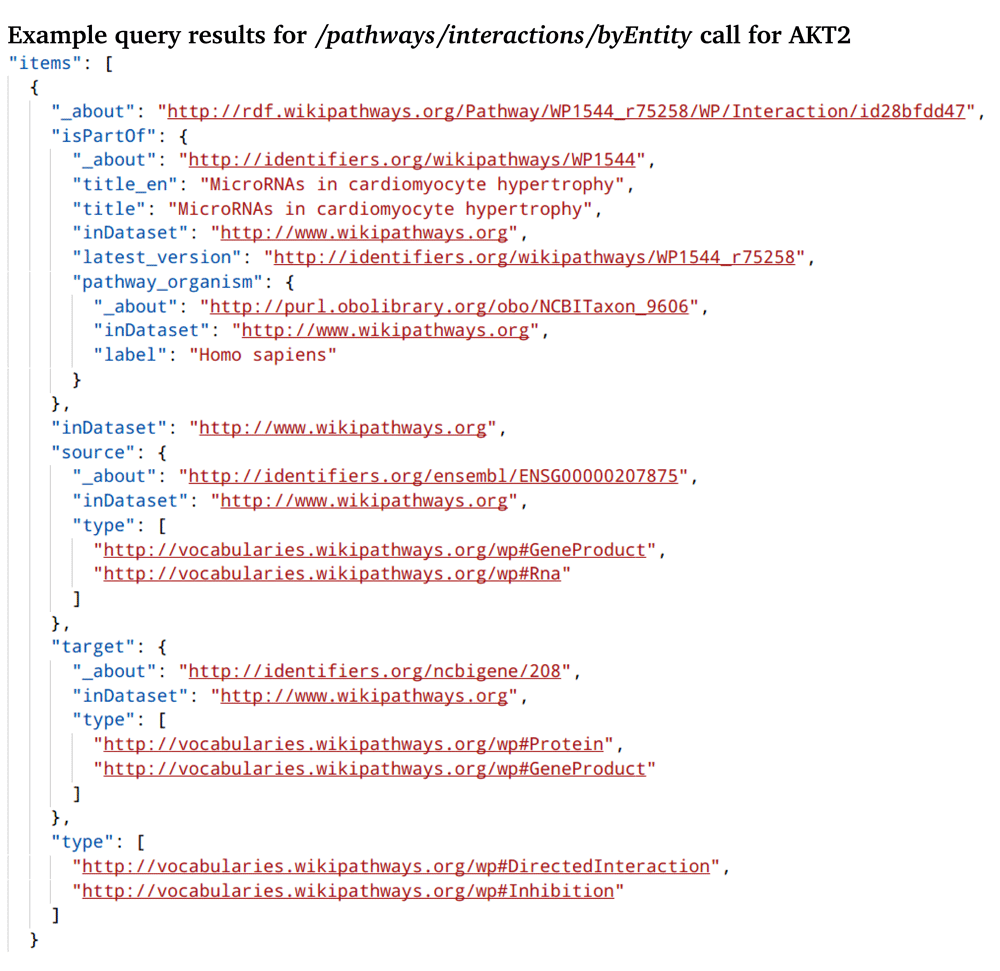
The participants of the interaction are directed from source (hsa-let7b) to target (AKT2). It also shows the type of interaction (inhibition), and the biological types of the interaction participants.
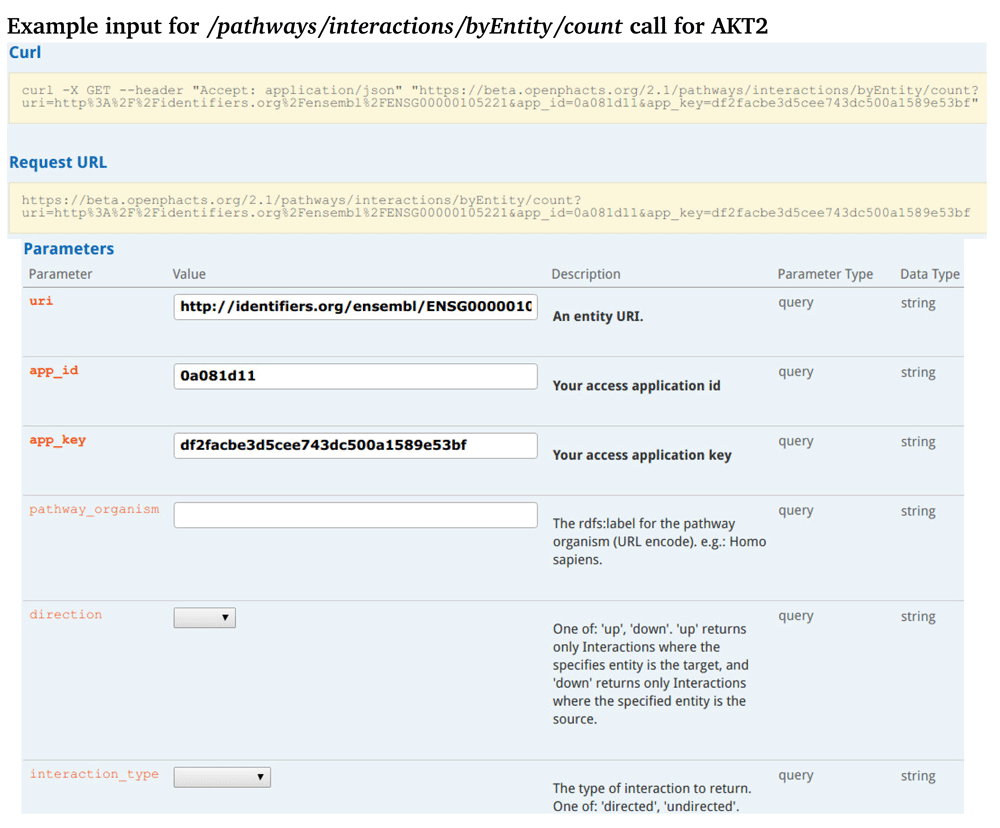
It takes a URI for an entity, in this case the Ensembl ID for AKT2 and returns a count of the interactions to which this gene product is involved. Only the entity URI, app ID, and app key are required fields. Optional parameters are pathway organism, direction, or type of interaction.
The other call implemented, /pathway/getInteractions (Figure 4), demonstrates an API call to return all interactions in the MicroRNAs in cardiomyocyte hypertrophy pathway15. This pathway has interaction details for AKT, mTOR, and PI3K, which are all important targets in cancer research16. For each interaction the participants are given and whether it is a directed or undirected interaction. An example result for this query can be seen in Supplementary Figure 2.
In order to demonstrate the basic use of the introduced API methods, we developed two workflows, available in the Supplementary Material. One uses Python to return a file with the results in a table and the other uses a HTML webpage using the ops.js JavaScript client library17. More involved workflows have been developed for KNIME and Pipeline Pilot18,19.
The Python script example uses the Open PHACTS /pathway/getInteraction API call and prompts the user to enter a WikiPathways pathway number that they wish to query, such as 1544 for WikiPathways pathway WP1544. Invocation of the API call with the pathway identifier returns information about the directed interactions that are involved with the pathway. The information that is returned is the interaction ID used by WikiPathways, the interaction type, and URIs for the source and target of the interaction. In order to convert the URIs into something more readable, a SPARQL query is then executed to get labels, from the WikiPathways SPARQL endpoint, for the source and target of the interaction. The results are written to a file with the interaction ID, interaction type, URIs for the source and target, as well as alias IDs, the curl for the API call, the pathway ID used, and a number of interactions returned.
The second example uses a HTML5 webpage and the ops.js JavaScript client library to retrieve interactions for a particular gene, using the URI for the gene’s Ensembl identifier and the /pathways/interactions/byEntity API method. The ops.js library passes the returned JSON with interaction information to a callback function, where the interacting source and target are extracted and the interacting entity determined. For each interacting entity, which may be a protein, RNA, or small compound, a call to the /pathways/interactions/byEntity/count method is made to return the number of interaction that entity has.
While the calls identified here are simple calls, workflow tools make it possible to take advantage of the integrative nature of the OPDP to make API calls in succession. Two such workflow tools that work with the OPDP are KNIME and Pipeline Pilot. With these tools, it is possible to perform a directional query of a target and identify alternative targets that can then be queried against the chemistry calls to identify active compounds for these alternative targets. The client libraries ops.js, ops4j, and ropenphacts also support Open PHACTS and the interaction calls for pathways. This allows users to perform API calls to the OPDP using their preferred language or platform, such as JavaScript, Java, or R.
The addition of interactions with direction information allows OPDP to answering more of the pre-defined scientific questions2. The directional information allows the user to explore how proteins and gene products are connected with one another and easily access this information. This is illustrated in the example queries using the cancer target AKT.
Online service: https://dev.openphacts.org/docs/2.1
Latest source code is available at: https://github.com/openphacts/OPS_LinkedDataApi
Archived source code of discussed version: https://doi.org/10.5281/zenodo.106825220
License: Apache License 2.0
This work was supported by the Innovative Medicines Initiative Joint Undertaking [grant number 115191], resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and EFPIA companies’ in kind contribution.
A special thanks goes to all members of the Open PHACTS project that provided the platform that was necessary.
Supplementary File 1. Contains two additional figures that show example API call output and contains the workflows to demonstrate the use of the APIs.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Partly
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Partly
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 12 Oct 18 |
read | read |
|
Version 1 17 Jan 18 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)