Keywords
MEG3 lncRNA, triple helix, triplex target sites (TTS)
MEG3 lncRNA, triple helix, triplex target sites (TTS)
Many human long noncoding RNAs are localized in the nucleus and potentially can participate in chromatin formation and remodeling1. Recently, technologies such as ChIRP2, ChRIP3, ChOP4, CHART5, RAP6, MARGI7 and GRID8 have been developed to map the genomic interacting sites of various lncRNAs. Although these techniques determine location of RNA binding sites, they are unable to clarify the interaction mechanisms. LncRNAs are capable of binding chromatin proteins, nascent RNA, single-stranded or double-stranded DNA, forming R-loops or triple helixes, respectively.
Maternally expressed gene 3 (MEG3) is one of the lncRNAs known to target chromatin. Genome-wide mapping of MEG3 with the chromatin oligo affinity precipitation followed by deep sequencing (ChOP-seq) method reveals that MEG3 binding sites are widespread, contain GA-rich motifs, and form RNA-DNA triplex structure4. Growing body of evidence shows that RNA-DNA triplex formation plays important role in RNA-chromatin interactions. Moreover, it has been shown earlier that triplex target sites (TTS) are frequently located near regulatory regions (including gene promoters) in the human genome9. In this work we investigate whether the DNA sites capable of triplex formation are specific enough to be regulated by a particular lncRNA.
After mapping 6837 ChOP-seq MEG3 peaks from hg19 to the hg38 using liftOver (the tool was downloaded from the UCSC Genome Browser on Nov 7, 2017 and ran as follows: liftOver MEG3.hg19_peaks.bed, hg19ToHg38.over.chain.gz MEG3.hg38_peaks.bed, unMapped.txt), 6694 peaks shorter than 1000 bp were used. Next, we selected 3kb regions (bins) centered at the peak middle positions using bedtools (version 2.25.0). The 3620 bins with overlapping genes (according to the GENCODE ver. 27 annotation10) were selected as true positives. Additionally, 3620 genomic regions of the same length and GC-content overlapping the GENCODE genes were randomly selected from the human genome as true negatives (the control bins). The validation set consisted of two subsets of bins without MEG3 peaks. Namely, another group of 3620 control bins were sampled and combined with the 3620 true negative regions from the test set.
We predicted triple helixes using Triplexator11 (version 1.3.2) with the settings recommended at the official website: -l 15 -e 20 -c 2 -fr off) with the following RNA queries:
- MEG3 (NR_002766: length = 1595 nt, GC-content = 57.55%),
- BE2L6 (NM_198183: 1620 nt, 57.59%),
- LILRA3 (NM_006865: 1608 nt, 57.71%),
- HMOX1 (NM_002133: 1590 nt, 57.80%).
The transcript sequences similar to the MEG3 in length and GC content were found using the RANN (version 2.5.1) R package12. UBE2L6, LILRA3 and HMOX1 were used to verify the sequence specificity of the MEG3-DNA hybridization. Additionally, the three random query sequences were obtained by mono-nucleotide shuffling the original MEG3 transcript using a custom Perl script. The Triplexator score for each genomic region was calculated as the sum of the scores of all the predictions between RNA and the genomic region.
The Triplexator tool11 was used to predict the RNA-DNA interactions between the MEG3 transcript and the 7240 genomic regions (bins) from the test set according to the Hoogsteen and reverse Hoogsteen base pairing rules. As anticipated, the triplex scores predicted for the MEG3 peak-containing bins were stronger than for the control bins – the median Triplexator SumScores were 48 and 25, respectively (p-value = 5.2e-100, see Figure 1a).
Strikingly, in all cases the SumScores produced by Triplexator for 3 other human RNAs (UBE2L6, LILRA3 and HMOX1) and the MEG3 peak-containing bins were also stronger than the scores for the control bins. Moreover, the statistical significances of the observed SumScore differences for two out of the three mRNAs were higher than for the MEG3 predictions (p-values = 0, 3.0e-24 and 1.4e-174, respectively – see Figure 1b). To find out whether it is a general property of the human transcripts or the identified MEG3-TTS have a tendency to form triplexes with any RNA in a nonspecific manner, the three random sequences were generated by the mono-nucleotide shuffling of the MEG3 transcript. Once again, the statistical significant differences between the two sets of bins were observed in all three cases (p-values = 1.0e-143, 5.8e-41 and 1.3e-33 – see Figure 1c).
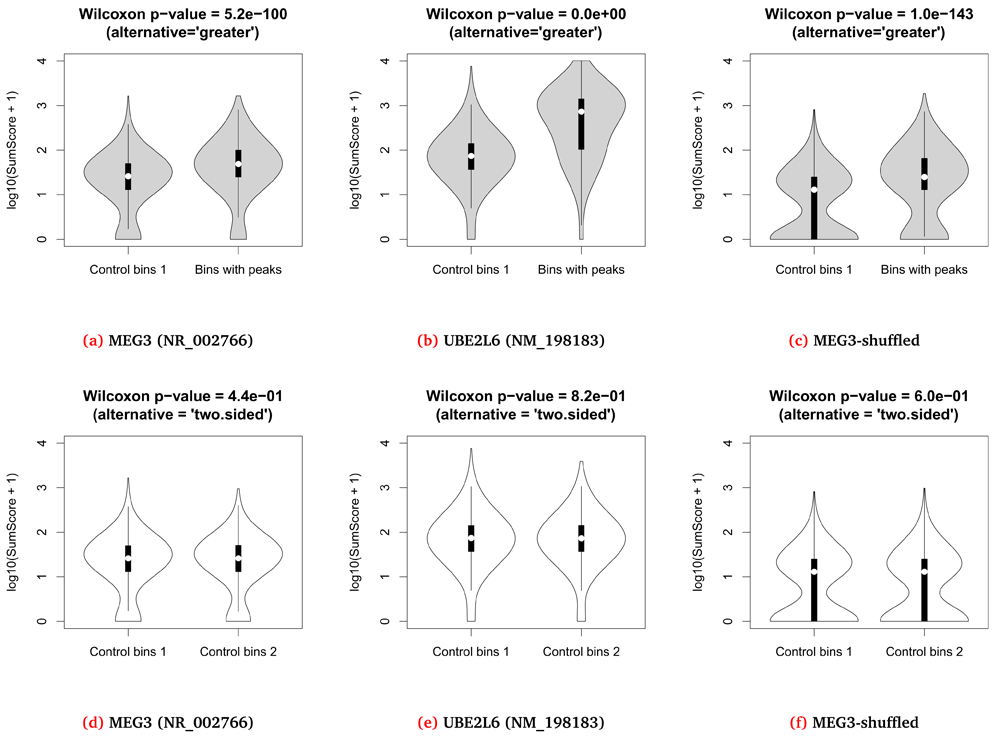
(a–c) The distributions of the Triplexator SumScores for the 3620 control regions without peaks and 3620 genomic regions with MEG3 peaks identified in the ChOP-seq experiment. (d–f) The distributions of the Triplexator SumScores for two sets of genomic regions without MEG3 peaks. The query transcript used in Triplexator run is indicated below each image.
To rule out a possibility of overprediction, we applied our computational approach to a ’validation set’ consisting of the MEG3 peak-free bins only (see Methods). On the contrary to the test set, no significant difference between the two groups of control bins was found for any of the seven RNA sequences (all p-values > 0.05, Figures 1d–f).
Our results suggest that at least in some cases, the formation of the RNA-DNA triplexes may be governed by the DNA sequence alone. If it is so, such ’universal’ TTSs are able to hybridize with various different RNAs almost irrespectively of their sequences (however the length and nucleotide content are probably important). Indeed, 18 peak-containing bins were present in the top 5% of the predictions for all seven tested RNAs. In contrast, there was only one such bin among the control regions. Notably, some of the 18 identified universal bins were extremely GA-rich (for example, hg38:chr5:93580373-93583373). The presence of the universal TTSs among the MEG3 peaks may explain the phenomena observed in our computational analysis.
Therefore, some parts of the human genome can hybridize with a number of different RNAs (or different regions of the same long RNA). It should be noted that one of the possible and actively discussed roles of the chromatin bound RNAs (including lncRNAs) is to bring different chromosomal parts together to enable the remote DNA-DNA interactions8. In the light of this biological role, the universal TTS can be viewed as the anchor point which can be bound by various nuclear RNAs to provide longdistance chromosomal contacts. Analysis of additional datasets is needed to further support this hypothesis.
Dataset 1: Coordinates of the original hg19-based and the converted hg38-based MEG3 peaks. DOI, 10.5256/f1000research.13522.d18975013
Dataset 2: Sequences of the seven queries as well as all the bins from the test and the validation sets. DOI, 10.5256/f1000research.13522.d18975114
Dataset 3: SumScores computed by running the Triplexator for each of the queries against the test or the validation set. DOI, 10.5256/f1000research.13522.d18975215
Dataset 4: [universal_TTS_bins.fna.gz]. Sequences of the bins containing putative ’universal’ TTSs. DOI, 10.5256/f1000research.13522.d18975316
This work was supported by the Russian Science Foundation [grant 14-15-30002].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We are thankful to Dr. Chandrasekhar Kanduri (University of Gothenburg, Sweden) for providing the original coordinates of the ChOP-seq peaks for the lncRNA MEG3.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
No
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genome analysis, lncRNA analysis, molecular evolution
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Mondal T, Subhash S, Vaid R, Enroth S, et al.: MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures.Nat Commun. 2015; 6: 7743 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
No
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
No
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Regulation of gene expression by noncoding RNA
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 09 May 19 |
read | read | |
|
Version 1 17 Jan 18 |
read | read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)