Keywords
Antibiotic resistance, Colistin resistance, Pan-drug resistance, Gram negative bacteria, Genomics, Bioinformatics
This article is included in the Pathogens gateway.
This article is included in the Antimicrobial Resistance collection.
Antibiotic resistance, Colistin resistance, Pan-drug resistance, Gram negative bacteria, Genomics, Bioinformatics
In the recent past, old antibiotic classes previously deemed unfit for treatment of bacterial infections due to associated toxicity concerns have been recommended for treatment in this type of infection1,2. This has been attributed to the emergence of resistance to the most recently considered last line antibiotics, the carbapenems1,2. Carbapenem resistance has been documented in bacteria belonging to the Enterobacteriaceae family, Acinetobacter baumannii and Pseudomonas aeruginosa1,2. The adoption of the old antibiotic agent category in routine empirical treatment has witnessed the use of a number of antibiotics such as colistin1,2.
Despite this reversion, gram-negative bacteria continue to undergo chromosomal mutations, which render their respective treatments virtually impossible and hence a major threat to global public health. The effects of these antibiotic resistance mutations are further exacerbated by horizontal transfer of antibiotic resistance genes in the same bacteria. As such, this paper explores the current documented trends of colistin resistance in several African settings. Additionally, it also describes the evolution of antibiotic resistance to plasmid mediated colistin resistance and the potential role of genomics and bioinformatics in precision antibiotic therapy targeted towards combating colistin resistance and antimicrobial resistance.
Data on the antimicrobial resistance burden, particularly colistin resistance, in Africa remains limited3. In 2014, the World Health Organization reported that antimicrobial resistance surveillance in Africa was a particularly difficult feat due to the scarcity of viable medical data, statistical information and unreliable laboratory capacity3. Despite this, African countries remain un-exempted from this worldwide antibiotic resistance trend that has emerged not only within hospital settings, but also disseminated within the community. The limited available literature from African settings has reported the mcr-1 gene mediated colistin resistance to be most prevalent in Africa, largely in South Africa, and this covers the largest portion of Africa according to the global map (Figure 1)4–7. Beyond South Africa, the emergence of colistin resistance has been reported in Algeria, Rwanda and Uganda8–10.
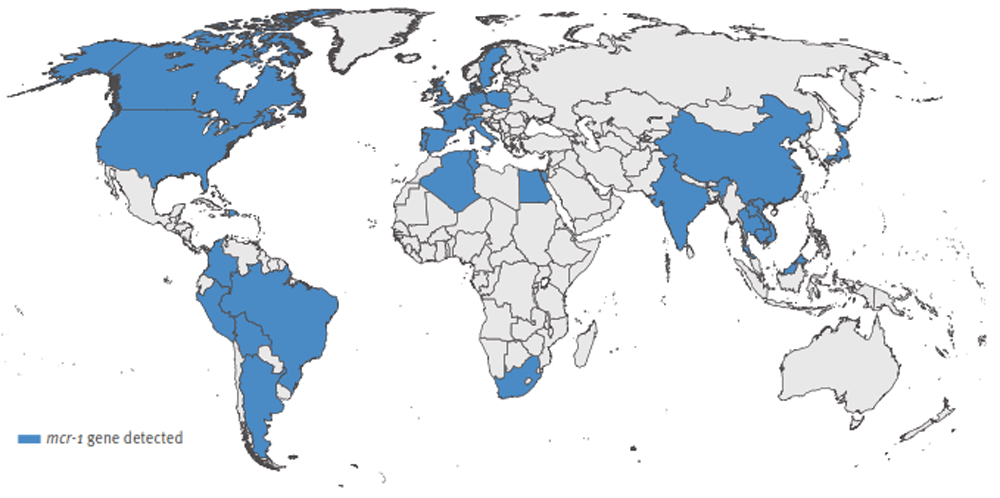
Adapted from Xavier et al.4 under a CC-BY 4.0 license.
Colistin (polymyxin E) is part of an old generation of antibiotics11 that form a family of cationic polypeptides. These are characterised by having a lipophilic fatty acyl side chain12–14. No exact mechanism of bacterial killing has been documented for polymyxins, especially in Acinetobacter spp14,15. However, a two-step mechanism has been described to elucidate their possible mechanism of action13,14.
The two stages involve: i) initial binding to and permeabilization of the outer membrane and ii) the destabilisation of the cytoplasmic membrane of the bacteria12–14. As a consequence, colistin functions by intercalating into the inner membrane following diffusion from the outer membrane across the periplasm and consequently causing the formation of pores, a phenomenon that results in bacterial lysis, which follows initial binding to bacterial surfaces12–14. Initial binding of colistin to the bacterial surface chiefly depends on the electrostatic interaction between the positively-charged colistin and the negatively charged phosphate group of lipid A, an endo toxic component on the lipopolysaccharide localised on the outer leaflet of the bacterial outer membrane12,14.
The modifications of the lipid A, which reduce and/or abolish the initial charge-based interaction with the polymyxins in bacteria13,15,16 and also the addition of either/or the 4-amino-4-deoxy-L-arabinose (L-Ara4N) and the phosphoethanolamine (PEtn) that ultimately form the basis of colistin resistance in bacteria16, is mediated by chromosomally encoded genes. These are involved in the modulation of two component regulatory systems; PmrA/PmrB and PhoP/PhoQ and mgrB, a negative regulator of the PhoP/PhoQ signalling system13–16.
Although initially thought that this resistance could not be spread from cell to cell (plasmid mediated)16, currently studies have shown otherwise. These have alluded transfer of colistin resistance among bacteria via plasmids in horizontal gene transfer4,16,17. Plasmid transfer of the colistin resistance mobile genes, mcr-1, mcr-2, and also mcr-34,16.
The treatment of infections involving antibiotic resistant gram-negative bacteria has become increasingly difficult overtime, a factor that has greatly contributed to high morbidity, mortality and high costs of health care18,19.
Currently, antibiotic resistance in these bacteria spans across several classes but likely follows a precise hierarchy of acquisition; this is mostly characterised by acquisition of “enhanced resistance” against more potent antibiotics following primary acquisition of “weaker resistance” against the less potent antibiotics alongside intrinsic resistance mechanisms in these bacteria, a trend that follows a Darwin’s like fashion20–23. These changes are a function of horizontal gene transfer, via conjugation, transformation and transduction24–27.
Resistance in gram-negative bacteria has been seen to transit from being mediated by the extended spectrum β-lactamases, a group of enzymes that can be disseminated among bacteria28,29; these chiefly confer resistance against broad spectrum cephalosporins. However, they also confer resistance to penicillins, monobactams and some carbapenems, particularly the Klebsiella pnemoniae carbapenemase, KPC28,30,31. In the same hierarchy are AmpC β-lactamases that form another group of β-lactamases, derived from older broad spectrum β-lactamases. These provide an even more extended activity that includes resistance against the cephamycins alongside resistance to penicillins, monobactams and cephalosporins32–34. These enzymes have in recent times been shown to not only be limited to being encoded on the chromosomes of bacteria, but have also been documented to have the potential of being disseminated via plasmids in horizontal gene transfer28,34,35 and also to co-exist with the extended spectrum β-lactamases36,37; factors that have made these bacteria “better resistant” to antibiotics. Next in the hierarchy are the carbapenemases, these enzymes are chiefly acquired in horizontal gene transfer and confer resistance to carbapenems alongside resistance to penicillins, broad spectrum cephalosporins including cefepime, a fourth generation cephalosporin, monobactams, aminoglycosides, quinolones and fluoroquinolones28,38. The development of resistance mediated by these enzymes to the different classes of antibiotics in these bacteria has been attributed to various factors among which is their use in therapy. This has not only abetted maintenance of resistance via selecting for resistance to these antibiotics in these bacteria but has also created a gap, a need for alternative antibiotics in therapy to replace the penicillins, β-lactams, carbapenems and the other classes of antibiotics used in the treatment of infections involving the drug resistant gram-negative bacteria4,16.
Colistin, a polypeptide antibiotic, a relatively old antibiotic, has been currently relied upon to provide the ultimate line of refuge against infections caused by antibiotic resistant gram-negative bacteria despite its previously documented impacts on health4,16. Colistin also appears to offer a choice in the face of almost no new antibiotics in production pipelines4,16,17.
Worryingly, the use of colistin is under threat due to the development of the novel plasmid mediated colistin resistance mediated by mcr-1, mcr-2 and mcr-3 heralds4,16. This provides a new challenge as bacteria that express these resistance genes assume the lead in the antibiotic resistance hierarchy and are distinctively extensive or worse pan drug resistant16,39–42.
Molecular studies previously done have reported colistin resistant gram-negative bacteria to also be resistant to an array of antibiotics. These bacteria have also been reported to carry plasmids that have been found to carry alongside colistin resistance genes, β-lactamases43,44, carbapenemase encoding genes45 and genes that code for resistances to other antibiotic classes that may include quinolones, fluoroquinolones and aminoglycosides13. Additionally, the carriage of mcr-1 has been documented as a possible indicator of resistance to the third generation cephalosporins and carbapenems38,44. Furthermore, these genes have been found to be co–carried with other resistance determinants in plasmids13,44,46,47; these genes represent a novel mechanism of antibiotic resistance in bacteria and a threat to the existing antibiotic therapy. Worsening the situation is the ability of selection for colistin resistance via the use of the extended spectrum cephalosporins. Additionally the use of tetracycline and sulphonamides has also been reported to contribute to the dissemination of colistin mobile gene carrying plasmids44,46. Also, worth noting is plasmids that carry colistin resistance genes have also been found to mostly carry other antibiotic resistant genes13,44–46.
Advances in technology including the rapidly growing field of genomics, are transforming clinical medicine48 and high-throughput sequencing technology (HTS) is increasingly being used in clinical microbiology49. HTS, with relatively simple bench top technology and efficient genomic library preparation protocols, has significantly improved the capacity to perform low-cost, efficient whole-genome sequencing (WGS), and has made it a feasible tool to enhance clinical diagnostic investigations in near real-time48. The processes generally involve culture-free parallel sequencing, producing vast quantities of genomic data that require modern computation techniques to assemble the genomic sequence reads as well as performing ensuing analyses that range from identifying the bacterial species or strain, antibiotic resistance mutations in the bacterial genomes, while ensuring the highest possible discriminatory power ever achieved by any technology49. Apart from this, WGS of bacteria can identify genes associated with virulence and pathogenicity as well as discover new genetic mechanisms for virulence, pathogenicity and antibiotic resistance48,50,51.
The identification and prediction of antibiotic resistant microorganisms in clinical specimens solely by molecular means in the diagnostic microbiology laboratory is not novel52. HTS technologies and computational tools offer unprecedented ability to sequence multitudes of bacterial genomes and enable interpretation of the resultant sequence information in near “real-time”52.
WGS represents the pinnacle for bacterial strain characterisation and epidemiological analyses. It is rapidly replacing traditional typing methods, antibiotic resistance gene detection and other molecular-based investigations in the near future. HTS technologies are rapidly evolving and their implementation in clinical and public health microbiology laboratories is increasing at a similar pace. These require standardised sample quality control, data interpretation, bioinformatics expertise, and infrastructure. The term ‘bioinformatics’ encompasses the handling and analysis of genomic sequence data, usually with the assistance of computer-based algorithms. Both ‘open source’ and commercially available bioinformatics programs/tools have been specifically developed for use in a clinical setting. However, many of practising healthcare workers in current practice have limited bioinformatics knowledge48.
Furthermore, phenomena such as genome plasticity and pan genomes that have the ability to influence bacterial resistome can only effectively be investigated using HTS and bioinformatics analyses. Understanding the bacterial genome dynamics is an important step in identifying the forces behind the observed antibiotic resistance and therefore be able to effectively manage the disease in question.
The bottleneck that remains in implementing WGS for clinical purposes is post-sequencing data analysis48.
Antibiotic resistance in bacteria is generally a natural phenomenon53–55 though augmented by human behaviour. Therefore, it is imperative to harness the best HTS technologies that sequence DNA at unprecedented speed, to enable previously unimaginable scientific achievements and novel biological applications56. Such applications of genomics tools has revolutionized microbial ecological studies and drastically expanded our view on the previously underappreciated microbial world57 including acquisition and transmission dynamics of antibiotic resistance. Single-Molecule Real-Time (SMRT) sequencing (Pacific Biosciences Inc.) in clinical microbiology has finally been realized at many levels in health care systems in the developing world and relatively only used during isolated scenarios of disease outbreaks in the less developed countries. These developments in HTS must be matched with continued efforts to improve the current bioinformatics analytic pipelines. Applying SMRT while genome sequencing to investigate bacterial colistin resistance would be made possible to predict resistance mutations, resistance mechanisms, trends, and patterns enabling efficient management of the colistin resistance by healthcare providers and pharmaceutical companies.
It is known that host, bacterial and environmental factors interact collectively to bring about antibiotic resistance. Therefore, HTS should be applied to a wide range of global collections of bacterial whole genomes to identify and predict new antibiotic drug resistance mutations using appropriate computational and bioinformatics algorithms.
Computational algorithms and tools offer ability to simulate bacterial genomic mutations while also offering possible clues on the mechanisms that may be shaping these mutations. These can as well be utilised to develop therapeutic interventions that may be used to target both the current and future acquired antibiotic drug resistance mutations.
No data are associated with this article
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the topic of the review discussed comprehensively in the context of the current literature?
Partly
Are all factual statements correct and adequately supported by citations?
Partly
Is the review written in accessible language?
No
Are the conclusions drawn appropriate in the context of the current research literature?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Antimicrobial resistance
Is the topic of the review discussed comprehensively in the context of the current literature?
Partly
Are all factual statements correct and adequately supported by citations?
Partly
Is the review written in accessible language?
Yes
Are the conclusions drawn appropriate in the context of the current research literature?
Partly
References
1. Hasman H, Hammerum AM, Hansen F, Hendriksen RS, et al.: Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015.Euro Surveill. 2015; 20 (49). PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular biology, bioinformatics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 20 May 19 |
read | read | read |
|
Version 1 04 Feb 19 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)