Keywords
DNA structure, Watson and Crick model, plasmid denaturation, single-stranded circular DNA, non-helical structure, side-by-side model.
At the request of the author, the article titled “Non-intertwined strands of plasmid DNA contradict the Watson and Crick model of DNA structure” [version 3;
... r review: 1 approved, 1 approved with reservations, 1 not approved]. F1000Research 2020, 8:356 (https://doi.org/10.12688/f1000research.18134.3) has been retracted from F1000Research. Previous versions of this article published on F1000Research have also been retracted (https://doi.org/10.12688/f1000research.18134.1; https://doi.org/10.12688/f1000research.18134.2).Since publication, Dr Pawan Kumar has performed additional experiments on the structure of a DNA molecule and was not able to generate further results that support this article. As such, they are no longer confident in the conclusions drawn in this article, and wish to retract. They apologise for any inconvenience caused.
At the request of the author, the article titled “Non-intertwined strands of plasmid DNA contradict the Watson and Crick model of DNA structure” [version 3; peer review: 1 approved, 1 approved with reservations, 1 not approved]. F1000Research 2020, 8:356 (https://doi.org/10.12688/f1000research.18134.3) has been retracted from F1000Research. Previous versions of this article published on F1000Research have also been retracted (https://doi.org/10.12688/f1000research.18134.1; https://doi.org/10.12688/f1000research.18134.2).
Since publication, Dr Pawan Kumar has performed additional experiments on the structure of a DNA molecule and was not able to generate further results that support this article. As such, they are no longer confident in the conclusions drawn in this article, and wish to retract. They apologise for any inconvenience caused.
DNA structure, Watson and Crick model, plasmid denaturation, single-stranded circular DNA, non-helical structure, side-by-side model.
DNA, Deoxyribonucleic acid
NaOH, Sodium hydroxide
HCl, Hydrochloric acid
hmP19, higher electrophoretic mobility band of pUC19 plasmid.
DNA is the genetic material of all organisms, with the exception of some viruses. The currently accepted model of DNA structure was proposed by James Watson and Francis Crick in 19531. According to this model, a DNA molecule consists of two antiparallel polynucleotide chains, intertwined with each other. Although the Watson and Crick (W/C) model is accepted widely, some researchers have raised questions against it and proposed alternative models for DNA structure2,3. Among these, Rodley’s model which envisaged that two strands of a DNA molecule are held side-by-side has generated significant interest and curiosity in the scientific community2.
In the present study, the W/C model of DNA structure was examined with the help of plasmid DNA. It was hypothesized that two strands of a plasmid will remain intertwined and not separate into single-stranded circular DNA molecules under denaturing conditions, if it follows the W/C model. To test this, pUC19 and pBR322 plasmids were denatured using sodium hydroxide (NaOH) and analyzed by gel electrophoresis. Interestingly, addition of NaOH to pUC19 and pBR322 plasmids resulted in new form of DNA showing higher electrophoretic mobility in agarose gel. DNA corresponding to higher electrophoretic mobility band of pUC19 (hmP19) was found to be single-stranded and circular, suggesting the separation of two strands of plasmid DNA. Under suitable conditions, hmP19 DNA re-annealed to form native pUC19 plasmid. These results showed that two strands of a DNA molecule are not intertwined with each other and contradicted the W/C model of DNA structure.
pUC19 and pBR322 plasmids were isolated from E. coli strains [cultured in Luria Bertani broth (HiMedia, catalogue# M1245) in a shaking incubator at 37°C] by alkaline-lysis method as described previously4. For denaturation, approximately 5 µg plasmid DNA (concentration determined using NanoDrop spectrophotometer) was added with an equal volume (5 µl) of NaOH solution of indicated concentration.
NaOH in denatured pUC19 solution was neutralized using 5 µl HCl (concentration, 0.5 M) and plasmid was incubated with Hind III (SibEnzyme, catalogue# E073), S1 nuclease (Promega Corporation, catalogue# E576A) or exonuclease I and alkaline phosphatase (Thermo Scientific, catalogue# EN0581 and EF0651, respectively) at room temperature for 30 min. Samples were immediately run on 1% agarose gel. A mix of forward primer (5’-CTGCTTTCCTGCATGTTC-3’) and reverse primer (5’-AAGCCCTTGCTTCTTATACT-3’) (experimental control) was also digested with exonuclease I and alkaline phosphatase. Digested and undigested primers were used in polymerase chain reaction [initial denaturation, 95°C/3 min followed by 30 cycles of (i) 95°C/30 sec, (ii) 55°C/30 sec, (iii) 72°C/30 in the same order] to amplify target sequence in A2780 cell line genomic DNA. PCR product was analyzed on 1% agarose gel.
hmP19 DNA bands were cut with the help of a clean knife. DNA was purified using an extraction kit, as suggested by manufacturer (FairBiotech, catalogue# DE0100). Briefly, gel was dissolved in DE buffer and passed through a column. After washing with buffers, DNA was eluted in 50 μl nuclease free water.
Transformation of E. coli strain DH5-alpha with gel-extracted plasmid DNA was carried out by heat-shock method (42°C for 30 sec) using water bath. Transformed and non-transformed bacteria were spread on ampicillin-nutrient agar plates supplemented with 25 µl X-Gal (Himedia, catalogue# MB0690. Plates were kept overnight in a 37°C incubator.
Two strands of a DNA molecule are held together by non-covalent interactions, which can be disrupted by increasing the pH of DNA solution5. In the present study, approximately 5 µg of plasmid DNA was denatured by adding NaOH solution of increasing concentration. It was observed that addition of 0.5 N NaOH to pUC19 resulted in a new form of DNA showing higher electrophoretic mobility in agarose gel (Figure 1a, Underlying data6). Similar results were obtained with pBR322 plasmid added with 0.5 N NaOH (Figure 1b, Underlying data7). Formation of higher electrophoretic mobility DNA in pBR322 plasmid added with NaOH has also been reported previously8.
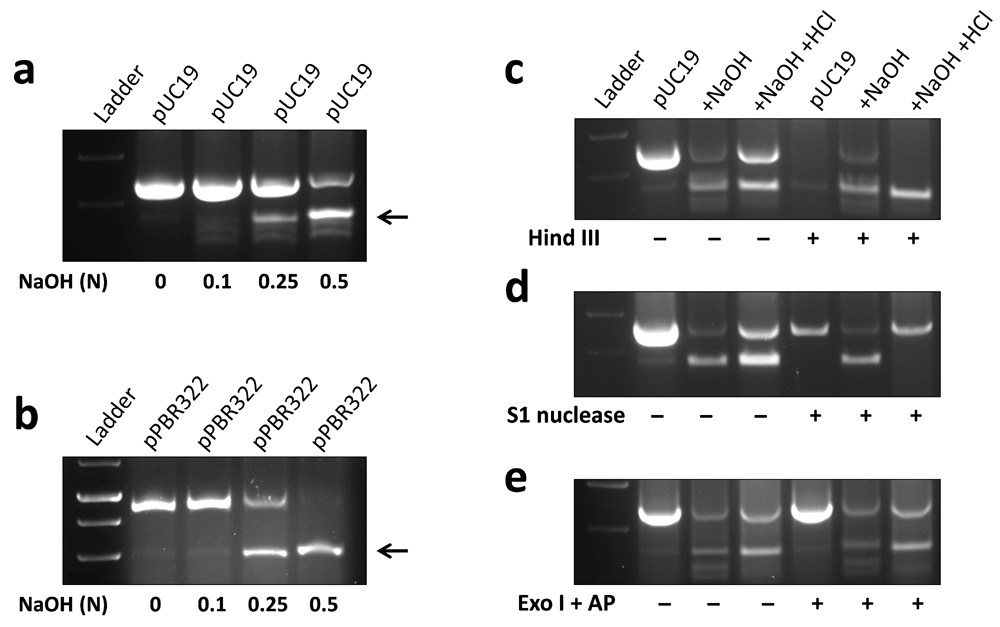
Approximately 5 µg plasmid DNA was added with an equal volume (5 µl) of NaOH solution of indicated concentration (normality, N) and run on 1% agarose gel. A distinct band of higher electrophoretic mobility DNA (marked by arrows) was observed in pUC19 (a) and pBR322 (b) plasmids added with 0.5 N NaOH. For enzymatic digestion, NaOH in denatured plasmid was neutralized using 0.5M HCl, followed by incubation with different enzymes. Native pUC19, but not hmP19 DNA was digested by Hind III, which acts on double-stranded DNA (c). S1 nuclease, which recognizes single-stranded DNA, degraded hmP19 DNA but not native pUC19 plasmid (d). Exonuclease I (Exo I) and alkaline phosphatase (AP), which eliminate single-stranded linear DNA, digested neither of native pUC19 or hmP19 DNA (e). Representative data of at least three independent experiments are shown.
DNA corresponding to higher electrophoretic mobility band of pUC19 (hmP19) was characterized using DNA modifying enzymes in next experiments. Incubation with Hind III, which acts on double-stranded DNA, digested pUC19 plasmid but not hmP19 DNA (Figure 1c, Underlying data9). S1 nuclease, which digests single-stranded DNA, degraded hmP19 DNA but not pUC19 plasmid (Figure 1d, Underlying data10). Exonuclease I and alkaline phosphatase, which would digest single-stranded linear DNA, degraded neither of pUC19 or hmP19 DNA (Figure 1e, Underlying data11). These results showed that hmP19 DNA is single-stranded and circular. Exonuclease I- and alkaline phosphatase-digested primers (experimental control) did not form product in PCR reaction (Supplementary Figure 1, Extended data12).
We asked whether hmP19 DNA was generated by separation of two strands of pUC19 or breakage of one strand followed by release of another one. Formation of a single band of higher electrophoretic mobility by denatured pUC19 (instead of two, which would have been the case when one strand was linearized and another was circular) suggested that hmP19 DNA was generated by separation of two strands of the plasmid (Figure 1a, b). To confirm this notion, hmP19 DNA was subjected to renaturing conditions. If hmP19 DNA were formed due to separation of two strands of pUC19, it would reanneal to form the native plasmid under suitable conditions. Interestingly, neutralization of NaOH with 0.25 M HCl resulted in the appearance of pUC19 plasmid in denatured DNA solution (Figure 2a, Underlying data13). To further confirm this, hmP19 DNA was extracted from gel and re-examined by gel electrophoresis. Both, gel-extracted hmP19 DNA and native plasmid DNA exhibited similar band patterns in the agarose gel (Figure 2b). These results showed that single-stranded circular hmP19 DNA was formed due to reversible separation of two strands of pUC19 plasmid DNA.
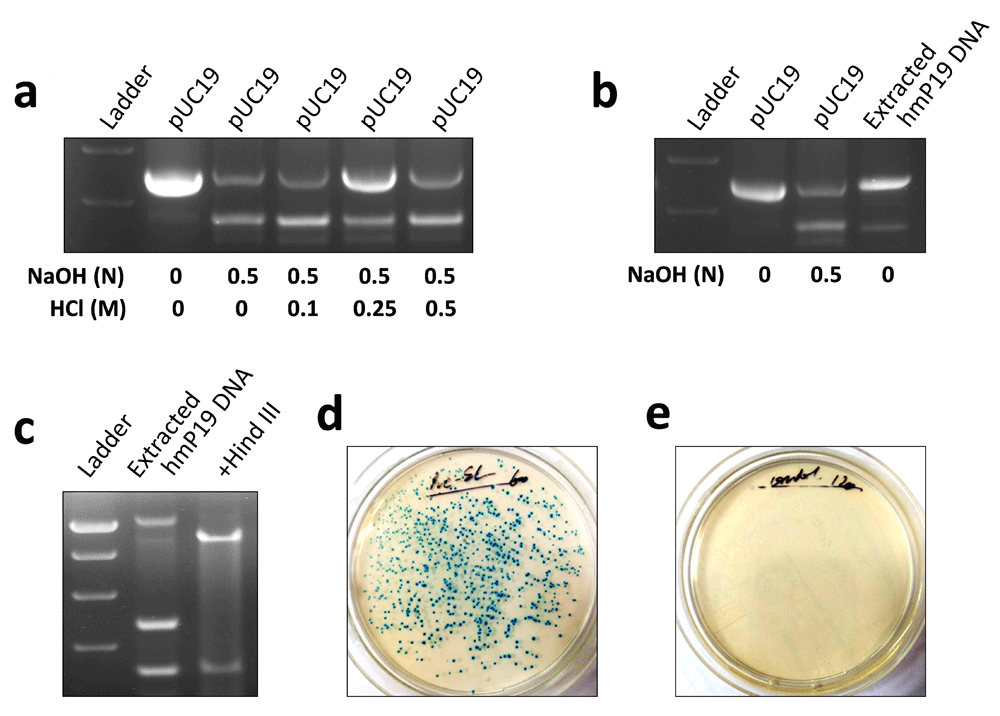
pH of denatured pUC19 plasmid DNA was normalized using HCl solution of indicated concentration. Agarose gel electrophoresis showed the formation of native pUC19 plasmid DNA in denatured plasmid DNA solution added with 0.25 M HCl (a). hmP19 DNA was extracted from agarose gel and approximately 3 µg of it was rerun on 1% agarose gel. Band pattern of pUC19 plasmid formed by extracted hmP19 DNA was same as that of native pUC19 plasmid (b). Gel-extracted hmP19 DNA was incubated with Hind III and run on 1% agarose gel. Hind III digested the pUC19 plasmid formed by gel-extracted hmP19 DNA (c). E. coli strain DH5-alpha was transformed with gel-extracted hmP19 DNA by heat-shock method. hmP19 DNA-transformed (d), but not non-transformed (e) bacteria formed colonies on ampicillin-X-Gal-nutrient agar plates. Representative data of two-three independent experiments are shown.
Next, we characterized pUC19 plasmid formed by reannealing of gel-extracted hmP19 DNA. Interestingly, similar to the native plasmid, pUC19 plasmid formed by gel-extracted hmP19 DNA was also degraded by Hind III (Figure 2c). Functionality of pUC19 formed by gel-extracted hmP19 DNA was demonstrated by its ability to transform E. coli. hmP19 DNA-transformed bacteria acquired ampicillin resistance and formed colonies on ampicillin-nutrient agar plates supplemented with X-Gal (Figure 2d, Underlying data14). No colonies were formed by non-transformed bacteria (Figure 2e). These results showed that pUC19 plasmid formed by re-annealing of hmP19 DNA was structurally and functionally similar to native plasmid DNA.
Concludingly, reversible separation of two strands of plasmid DNA into single-stranded circular DNA molecules shows that DNA strands are not intertwined with each other. These findings contradict the W/C model of DNA structure and provide evidence for the side-by-side structure of DNA.
Figshare: Addition of sodium hydroxide (NaOH) to pUC19 plasmid resulted in a distinct band of higher electrophoretic mobility DNA. https://doi.org/10.6084/m9.figshare.7751093.v66
This project contains the following underlying data:
Figshare: Addition of sodium hydroxide (NaOH) to pBR322 plasmid resulted in a distinct band of higher electrophoretic mobility DNA. https://doi.org/10.6084/m9.figshare.7751084.v57
This project contains the following underlying data:
Figshare: Higher electrophoretic mobility band of pUC19 (hmP19) DNA is single-stranded in nature. https://doi.org/10.6084/m9.figshare.7751090.v310
This project contains the following underlying data:
Figshare: Higher electrophoretic mobility band of pUC19 (hmP19) DNA is not double-stranded in nature. https://doi.org/10.6084/m9.figshare.7751087.v39
This project contains the following underlying data:
Figshare: Higher electrophoretic mobility band of pUC19 (hmP19) DNA is single-stranded and circular in nature. https://doi.org/10.6084/m9.figshare.7751081.v311
This project contains the following underlying data:
Figshare: pUC19 DNA formed by higher electrophoretic mobility band of pUC19 (hmP19) DNA demonstrates structural and functional properties of native plasmid14. https://doi.org/10.6084/m9.figshare.7751078
This project contains the following underlying data:
puc str fun.tif (Gel image showing plasmid products following digestion with Hind III and S1 nuclease, and plate images of bacteria transformed with purified hmP19)
Figshare: Single-stranded, circular higher electrophoretic mobility band of pUC19 (hmP19) DNA formed native double-stranded pUC19 plasmid with neutralization of sodium hydroxide (NaOH). https://doi.org/10.6084/m9.figshare.7754375.v113
This project contains the following underlying data:
Figshare: Supplementary Figure 1: Exonuclease I- and alkaline phosphatase-digested primers did not form product in polymerase chain reaction (PCR). https://doi.org/10.6084/m9.figshare.7754375.v112
This project contains the following extended data:
Supplimentary_Figure_1.jpg (Gel image showing digested primer products following incubation with exonuclease 1 and alkaline phosphatase)
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 3 (revision) 10 Nov 20 |
|||
|
Version 2 (revision) 01 Oct 20 |
read | ||
|
Version 1 01 Apr 19 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
- The higher electrophoretic mobility band of plasmids (hmpUC19 or hmpBR322) is actually random coils of the NaOH denatured plasmids composed of a pair of single stranded circular DNAs (ssc DNA). In alkaline solution, the denatured plasmids are paired ssc DNAs; they are probably tightly tangled with each other just without hydrogen bonds. In 0.25 N NaOH, the backbone of the plasmid is stable and keeps intact, however, due to topological rule, the paired ssc DNAs cannot separate from each other; they just tangled with each other. As soon as the paired ssc DNAs entered into agarose during agarose gel electrophoresis (AGE), the sudden pH change pushes the denatured ssc DNAs renature quickly but not in legitimate way i.e., adopting many inter-strand or intra-strand hydrogen bonds between the AT or GC pairs, causing the formation of tightly tangled entity with electric mobility higher than their supercoiled counterparts. The differences of the denatured plasmid in solution and in agarose gel are not commonly noticed by many scientists. It is reasonable and unquestionable that the NaOH denatured plasmids can renatured under suitable conditions as the author has indicated. It does not mean the alkaline denatured complementary ssc DNAs has been completely physically separated.
- If the phenomenon of higher mobility plasmid is really separated ssc DNAs, as the author supposed to be, they should be separable by AGE. The figure 3 in the report of BBA (Xu, YC. 2008. Finding of a zero linking number topoisomer. 2009. B.B.A. 1790, 126-133.) indicated that the two complementary plasmid strands of ssc DNA or ssl DNA of a singly nicked plasmid can all be separated on agarose gel. However, there is no indication that the two ssc DNA of NaOH denatured plasmids can be separated by AGE.
- If the phenomenon of higher mobility plasmid is really separated ssc DNAs, as the author supposed to be, their electrophoresis behavior should not be affected by the supercoiling of the plasmids. The figure 7 of a paper (Xu, YC, 2011, Replication Demands an Amendment of the Double Helix. In book: (Seligmann, H., Ed., DNA Replication-Current Advances, InTech, Rijeka, 29-56.) indicated that the mobility of denatured plasmid is closely related to the supercoiling of plasmid, the alkaline denature relaxed plasmids moves much faster than that of their highly supercoiled counterparts.
- All native plasmids are composed of a set of covalently closed circular DNA (ccc DNA), the author cannot provide a reasonable explanation on how the paired ssc DNAs overcome the topological rule without the help of strand passing ability of topoisomerases.
What the DNA really is? Many different answers can be heard from different observers. Just as a cubic or cylinder shaped iceberg seen by observers cannot jump to the conclusion that bottom of the iceberg is in the same shape.The DNA structure has been studied by many scientists all over the world for many decades. Although many facts and experimental results contradicted or collided with one of the claims of the Watson-Crick Model, it is still hard to know what DNA really is. Based on the ambidextrous double helix model, it is predicted that in any plasmid there is a zero linking number topoisomer. This double helix conjecture can be proved by experiment. It is not hard to do. (Xu, YC. 2019. Evidence falsifying the double helix model. Symmetry, 11, 1445).
Once the double helix conjecture was proven, it would be a shock to the molecular biology because it confirmed the prediction of the ambidextrous model is correct and its meaning can be easily understood by anybody with normal IQ. Therefore, it may induce a "Paradigm shift" and maintain lasting influence on the tertiary structure of DNA. This meme will be transferred generations after generations since knowledge cannot inherit from parents; every student has to learn from the simplest beginning. As textbooks are the main source of their knowledge, it would be guilty if we keep on teaching them the Watson-Crick Model that we know is wrong or questionable.
- The higher electrophoretic mobility band of plasmids (hmpUC19 or hmpBR322) is actually random coils of the NaOH denatured plasmids composed of a pair of single stranded circular DNAs (ssc DNA). In alkaline solution, the denatured plasmids are paired ssc DNAs; they are probably tightly tangled with each other just without hydrogen bonds. In 0.25 N NaOH, the backbone of the plasmid is stable and keeps intact, however, due to topological rule, the paired ssc DNAs cannot separate from each other; they just tangled with each other. As soon as the paired ssc DNAs entered into agarose during agarose gel electrophoresis (AGE), the sudden pH change pushes the denatured ssc DNAs renature quickly but not in legitimate way i.e., adopting many inter-strand or intra-strand hydrogen bonds between the AT or GC pairs, causing the formation of tightly tangled entity with electric mobility higher than their supercoiled counterparts. The differences of the denatured plasmid in solution and in agarose gel are not commonly noticed by many scientists. It is reasonable and unquestionable that the NaOH denatured plasmids can renatured under suitable conditions as the author has indicated. It does not mean the alkaline denatured complementary ssc DNAs has been completely physically separated.
- If the phenomenon of higher mobility plasmid is really separated ssc DNAs, as the author supposed to be, they should be separable by AGE. The figure 3 in the report of BBA (Xu, YC. 2008. Finding of a zero linking number topoisomer. 2009. B.B.A. 1790, 126-133.) indicated that the two complementary plasmid strands of ssc DNA or ssl DNA of a singly nicked plasmid can all be separated on agarose gel. However, there is no indication that the two ssc DNA of NaOH denatured plasmids can be separated by AGE.
- If the phenomenon of higher mobility plasmid is really separated ssc DNAs, as the author supposed to be, their electrophoresis behavior should not be affected by the supercoiling of the plasmids. The figure 7 of a paper (Xu, YC, 2011, Replication Demands an Amendment of the Double Helix. In book: (Seligmann, H., Ed., DNA Replication-Current Advances, InTech, Rijeka, 29-56.) indicated that the mobility of denatured plasmid is closely related to the supercoiling of plasmid, the alkaline denature relaxed plasmids moves much faster than that of their highly supercoiled counterparts.
- All native plasmids are composed of a set of covalently closed circular DNA (ccc DNA), the author cannot provide a reasonable explanation on how the paired ssc DNAs overcome the topological rule without the help of strand passing ability of topoisomerases.
What the DNA really is? Many different answers can be heard from different observers. Just as a cubic or cylinder shaped iceberg seen by observers cannot jump to the conclusion that bottom of the iceberg is in the same shape.The DNA structure has been studied by many scientists all over the world for many decades. Although many facts and experimental results contradicted or collided with one of the claims of the Watson-Crick Model, it is still hard to know what DNA really is. Based on the ambidextrous double helix model, it is predicted that in any plasmid there is a zero linking number topoisomer. This double helix conjecture can be proved by experiment. It is not hard to do. (Xu, YC. 2019. Evidence falsifying the double helix model. Symmetry, 11, 1445).
Once the double helix conjecture was proven, it would be a shock to the molecular biology because it confirmed the prediction of the ambidextrous model is correct and its meaning can be easily understood by anybody with normal IQ. Therefore, it may induce a "Paradigm shift" and maintain lasting influence on the tertiary structure of DNA. This meme will be transferred generations after generations since knowledge cannot inherit from parents; every student has to learn from the simplest beginning. As textbooks are the main source of their knowledge, it would be guilty if we keep on teaching them the Watson-Crick Model that we know is wrong or questionable.