Keywords
evolution, gene networks, fitness landscape learning
evolution, gene networks, fitness landscape learning
A central idea of modern biology is that evolution proceeds by mutation and selection. This process may be represented as a walk in a fitness landscape leading to fitness increase and slow adaptation 1 . According to classical ideas this walk can be considered a sequence of small random steps with small phenotypic effects. Nevertheless, there is a limited amount of experimental support for this idea 2 and some experimental evidence that evolution can involve genetic changes of a relatively large effect and that the total number of changes are surprisingly small 3 . Another intriguing fact is that organisms are capable of making adaptive predictions of environmental changes 4 .
To explain those facts new evolutionary concepts have been suggested (see the review by 5 and references therein). The main idea is that a population can “learn” (recognize) fitness landscapes 5– 7 . The approach developed in these works is a generalization of ideas from machine learning in which learning (regression to data) is viewed as selection and generalization (interpolation or extrapolation) is viewed as adaptation.
A mathematical basis for investigation of evolution learning problems has been developed by 8. However, this work uses a simplified model, where organisms are represented as Boolean circuits seeking an “ideal answer” to environmental challenges. These circuits involve N g Boolean variables that can be interpreted as genes, and the ideal circuit answer maximizes the fitness. A similar model was studied numerically by 7 to confirm the theory of “facilitated variation” explaining the appearance of genetic variations which can lead to large phenotypic ones. In the work by 9 a theory of the evolution of these Boolean circuits was advanced. It was shown that, under some conditions—weak selection, see 10—a polynomially large population over polynomially many generations (polynomial in N g ) will end up almost surely consisting exclusively of assignments, which satisfy all constraints. This theorem can shed light on the problem of the evolution of complex adaptations since that satisfiability problem can be considered as a rough mathematical model of adaptation to many constraints.
In 6 it is shown that, in the regime of weak selection, population evolution can be described by the multiplicative weight update algorithm (MWUA), which is a powerful tool, well known in theoretical computer science and a generalization of such famous algorithms as Adaboost and others 11 . Note that in 6 infinitely large populations are investigated whereas the results of 9 hold only for finite populations and take into account genetic drift.
Evolution Computation(EC) problems are considered recently by many papers 12– 16 mainly for artificial test fitness functions like OneMax or LeadingOnes (for an overwiew of EC problems, see 17).
In this paper, we investigate adaptation and the fitness landscape learning problem for more realistic fitness function This fitness can model adaptation for insects and connected with a fundamental hard combinatorial problem: K-SAT.
The main results can be outlined as follows. We show that, in a fixed environment, genes can serve as learners in the machine learning sense. Indeed, if an organism has survived for a long period, this fact alone constitutes important information, which can be used. The biological interpretation of this fact is simple: if a population is large enough and mutations are sufficiently rare, deleterious mutations are eliminated by purifying selection. Hence, those non-neutral mutant alleles which have become fixed in natural populations will, with probability close to 1, be adaptive and cause a positive increment of fitness (see Theorem 3.1 and Theorem 3.2 in Subsection 3.1 and Subsection 3.2). We obtain mathematical results, which allows us to estimate the reduction of mutation number due to that learning landscape procedure. Learning can sharply reduce the number of mutations needed to form a phenotypic trait useful for adaptation that is consistent with experimental data mentioned above (see 3.
Another important result is as follows. We estimate the accuracy of fundamental Nagylaki equations 6, 10 for a realistic population model, where the population size is bounded and a non-zero mutation rate is taken into account (in the case of asexual reproduction). Those accuracy estimates are fulfilled for all possible values of mutation rates and population sizes.
In this section, we describe our model and mathematical approach.
We assume that the genotype can be described by Boolean strings of length N g , where N g is the number of genes. Then
where s i = 1 means that gene i is activated (switched on) and s i = 0 means that it is repressed (switched off). Correspondence between Boolean hypercube and genotypes is considered for example in 12.
Although phenotype is controlled by genes, it is also influenced by environmental conditions and various epigenetic processes. In this paper, we suppose that phenotypic traits are controlled by genotype only. We consider levels f j of expressions of those traits as real variables in the interval (0, 1). Then the vector f = ( f 1, . . . , f N b ) can be considered to represent the organismal phenotype. We suppose that
where f j ∈ (0, 1) is a real valued function of the Boolean string s, the genotype.
Only a part of s i is involved in f j . Namely, for each j we have a set of indices K j = { i 1, i 2, . . ., i n j } such that f j depends on s i with i ∈ K j , so that
where i l ∈ K j and n j is the number of genes involved in the control of the trait expression. The representation of phenotype by the quantities f j is suggestive of quantitative traits because the f j are real valued. The limiting values of 0 or 1 suggest another interpretation, however, in terms of cell type. Multicellular organisms consist of cells of different types. One can suppose that the organismal phenotype is defined completely by the corresponding cell pattern. The cell type j is determined by morphogenes, which can be identified as gene products or signaling molecules that can change cell type (or genes that code for signaling molecules that can determine cell types or cell-cell interactions and then finally the cell pattern). The morphogene activity is defined by ( 2.2).
We further suppose:
Assumption M. Assume activities f j have the following properties.
The sets K j are independent uniformly random subsets of S g = {1, . . . N g }
We denote the total number of genes involved in regulation of all f j by N r , where
Assumption M implies that the genetic control of the phenotype is organized, in a sense, randomly, and that only a portion of the full set of genes controls phenotypic traits. That modularity of gene control is well known from experimental data (see 18, 19) and for evolution computation problems it was studied, for example, in 16.
Consider an example, where the assumption M holds, where we have a saturated expression, inspired by earlier work 20, 21 . Let
where j = 1, . . . , N b . Here σ( z) is a sigmoidal function of real z such that
and w ij , h j are some coefficients (their meaning will be explained below). As an example, we can take σ( S) = (1 + exp( −bS)) −1, where b > 0 is a sharpness parameter. Note that for large b this sigmoidal function tends to the step function and for b = + ∞ our model becomes a Boolean one. The parameters h j defines thresholds for trait expression 20 . The relation ( 2.4) can be interpreted as a simple mathematical model for quantitative trait locus (QTL) action.
To understand the role of h j consider a trait f j and suppose that for a well adapted organism f j ≈ 1. Let, for simplicity, w ij take the values 1, 0, or −1. Then the parameter h j defines how many genes involved in the control of the f j expression should be activators and how many should be repressors. Let the numbers of activator and repressor genes be respectively.
Then f j ≈ 1 if
One can suppose that h j describes a direct influence of environment on phenotype, such as stress, that can exert epigenetic effects. In Section 2.5 using data from 3 we will show that the model defined by ( 2.4) are capable to describe main topological characteristics of really observed fitness functions in the case of mimicry, camouflage and thermoregulation for insects.
Let us introduce the matrix W of size N b × N g with the entries w ij . The coefficients w ji determine the effects of terminal differentiation genes (see 23), and hence encodes the genotype-phenotype map. We assume that the coefficients w ji are random, with the probability that w ji > 0 or that w ji < 0 is β/2 N, where β > 0 is a parameter. This quantity β << N defines a genetic redundancy, i.e., averaged numbers of genes that control a trait. Note that then large β ≫ 1 one has n j < 4 β with the probability Pr β , which is exponentially close to 1: Pr β > 1 – exp(−0.1 β), thus, the number n j are bounded.
We know little about the details of how fitness relates to the phenotype of multicellular organisms, and for that reason classic neo-Darwinian theory takes fitness to be a function of genotype. Some models which take account of epistasis have been proposed 24 . The random field models assign fitness values to genotypes independently from a fixed probability distribution. They are close to mutation selection models introduced by 25, and can be named House of Cards (HoC) model. The best known model of this kind is the NK model introduced by Kauffman and Weinberger 26 , where each locus interacts with K other loci. Rough Mount Fuji (RMF) models are obtained by combining a random HoC landscape with an additive landscape models 27 . In evolution computations (EC) some artificial fitness models were used, for example OneMax and Leading Ones to test evolution algorithms, see for example 15 .
In this work, we use the classical approach of R. Fisher by introducing an explicit representation of phenotype, f, and allow it to determine fitness through interaction with an environment b. That is, we assume that the phenotype is completely determined by the phenotype trait expression, and thus the fitness depends on the genotype s via f j .
We express the relative fitness F and its dependence on environment b via an auxiliary function W via the relation
where K F is a positive constant and b = ( b 1,..., b N b ) is a vector consisting of coefficients b j , respectively. Below we refer to W as a fitness potential, and we assume that
Sometimes, if the parameter b is fixed, we shall omit the corresponding argument in notation for W and F.
We consider fitness as a numerical measure of interactions between the phenotype and an environment. For a fixed environment, this idea gives us the fitness of classical population genetics. A part of the fitness, however, depends on the organism developing properly and for now we represent it as independent of the environment, although we are aware that this is not always the case. Note that some coefficients b j may be negative and others may be positive, and that the model ( 2.7) can describe gene epistatic effects via dependence of f j on s if f j are nonlinear in s.
The expression ( 2.7) can serve as a rough approximation of the fitness function in the case of insects such as grasshoppers or fruit flies. In fact, important factors, which determine insect survival, are thermoregulation, mimicry and camouflage levels 18, 22, 28 . All those factors depend on colour pigmentation pattern. Blackwhite pigmentation patterns can be roughly described by vectors f = ( f 1, f 2,..., f N b ), where f j ≈ 1 and f j ≈ 0 mean that the cell j is black, or white, respectively, Then thermoregulation depends on The mimicry level can be approximately defined by expression , where f * is a target pattern corresponding to an insect to mimic. Colour patterns can be also described by classical RGB formalism.
The representation of the fitness as a sum of terms ( 2.7) is of course a rough approximation; however if assumption M holds that representation is consistent with important observed facts. First, mutations have been identified that alter one part of the pigment pattern without affecting any other. This independence of different pattern parts can be explained by the modular organization of the genetic regulation that controls pigmentation. In the course of evolution, different aspects of the pigment pattern have clearly evolved independently of each other 18 . Second, the topology of the fitness landscapes was studied in 22 by field experiments in the case of insect mimicry. Main conclusions are as follows. A number of studies of fitness landscapes in natural populations have demonstrated low fitness of intermediate phenotypes, i.e., existence of valley in the fitness landscape. It is found 22 that natural selection promotes genetic architecture preventing the expression of intermediate phenotypes. Close fitness peaks are separated by ridges, favouring colour pattern switches and allowing drift from local peaks.
In Section 2.5 we will show that the fitness model defined by ( 2.4) and ( 2.7) have those topological properties.
For simplicity, we consider populations with asexual reproduction. (Although a part of the results remain valid for sexual reproduction, as we discuss at the end of this subsection). We choose initial genotypes randomly from a gene pool and assign them to organisms. This choice is invariant with respect to the population member, i.e,. the probability to assign a given genotype s to a member of the population does not depend on that member.
In each generation, there are N pop( t) individuals, the genome of each of which is denoted by s( t), where t = 0, 1, 2,... stands for the evolution step number). Following the classical Wright-Fisher ideas, we suppose that generations do not overlap. In each generation (i.e., for each t), the following three steps are performed:
1. Each individual s at each evolution step can mutate with probability p mut per gene;
2. At evolution step t each individual with a genotype s produces k progeny, where k is a random non-negative integer, distributed according to the Poisson law
where q = F( s) is the fitness of that individual;
3. To take into account ecological restrictions on the population size, we introduce the maximal population size N popmax . If ( t) > N popmax, where ( t) is the number of progeny produced by the population at step t, we kill randomly selected individuals in a population-dependent manner. The probability of the death of an individual is given by p kill( ) = 1 − ( N popmax/ ( t)). If ( t) ≤ N popmax, we do nothing. We refer to this as the “massacre procedure.”
Conditions 1 and 2 imply that mutations in the genotypes create a new genetic pool and then a new round of selection starts. Condition 3 expresses the fundamental ecological limitation that all environments can only support populations of a limited size. If N popmax ≫ 1 then by ( 2.8) and the Central Limit Theorem one can show, under some additional conditions, (see Section 4) that fluctuations of the population size are small, and thus the population is ecologically stable and N pop( t) ≈ N popmax.
In the limit case of infinitely large populations we will write the discrete dynamical equation for the time evolution of the frequency X( s, t) of the genotype s in the population as
where ( t) is the average fitness of the population at the moment t defined by
where S( t) is the set of genotypes existing in the population at time t (the genetic pool) and X( s, t) = N( s, t) /N pop( t) is the frequency of the genotype s. Here N( s, t) denotes the number of the population members with the genotype s at the step t.
The equations ( 2.9) do not take mutations into account. They describe only changes in the genotype frequencies because of selection at the t-th time step. The same equations govern evolution in the case of sexual reproduction in the limit of weak selection 6, 10 . Note that for an evolution defined by ( 2.9), the average fitness ( t) defined by ( 2.10) satisfies Fisher’s theorem, so that this function increases at each time step t: ( t + 1) ≥ ( t).
Adaptation (i.e., maximization of fitness in a changing environment) is a very hard problem since over evolutionary history we observe the coevolution of many traits accompanied by changes in many genes. In its general context, this is a problem in the theory of macroevolution, which in general requires the integration of population genetics and developmental biology for its full understanding. There are two key components of this problem. First, development is itself a dynamical process operating over time. Second, there is a combinatorial component of development wherein different combinations of gene must be expressed in different cell types. This combinatorial aspect of the problem means that straightforward theoretical methods of considering the relationship between gene expression and a changing environment that have been very successful in single celled organisms 29 cannot be applied to metazoa. In this work, for the sake of tractability, we focus on the combinatorial aspect of the problem and neglect developmental dynamics. Even at the highly simplified level of our model, adaptation is a hard computational problem, as we now demonstrate.
Consider the case, where f j are defined by relations ( 2.4) and assume that
As a consequence of the second assumption, F attains its maximum for f 1 = 1, f 2 = 1,..., f N b = 1. Let us show that, even in this particular case, the problem of the fitness maximization with respect to s is very complex. In fact, for a choice of h j it reduces to the famous NP-complete problem, so-called K-SAT, which has received a great deal of attention in the last few decades (see 30– 35). The K-SAT can be formulated as follows.
K -SAT problem. Let us consider the set V n = { s 1,..., s n } of Boolean variables s i ∈ {0, 1} and a set m of m clauses. The clauses C j are disjunctions (logical ORs) involving K literals z i 1 , z i 2 ,..., z i k , where each z i is either s i or the negation i of s i. The problem is to test whether one can satisfy all of the clauses by an assignment of Boolean variables.
Cook and Levin 30, 31 have shown that the K-SAT problem is NP-complete and therefore in general it is not feasible in a reasonable running time. In subsequent studies—for instance, by 32 —it was shown that K-SAT of a random structure is feasible under the condition that N b < α c ( K) N g , where α c ( K) ≈ 2 K log 2 for large K.
The set K of solutions of random K-SAT has a nontrivial structure depending on parameter α = N b/N g 33, 35 . For sufficiently small α < α g ( K), where α g ( K) ≈ 2 K log( K) /K is some critical parameter, the set K forms a giant cluster, where nearest solutions are connected by a single flip and one can go from a solution to another by a sequence of single flips (pointed mutations) 35 . For α ∈ ( α g , α d ), where α d ( k) < α c is another critical value, solutions form a set of disconnected clusters. The local search algorithms do not work in the domain α > α g .
Probably, for evolution context K-SAT was applied first in 36, where it was used for an investigation of speciation problem.
To see the connection of our model with K-SAT, consider equation ( 2.4) supposing that w ij ∈ {1, 0, −1} and h j = −C j + 0.5, where C j is the number of negative w ji in the sum We set m = N b and n = N g . Under this choice of h j , the terms σ( S j ) can be represented as disjunctions of literals z j . Each literal z j equals either s j or j , where j denotes negation of s j . To maximize the fitness, we must assign s j such that all disjunctions will be satisfied. If we fix the number n j of the literals participating in each disjunction (clause) and set n j = K, this assignment problem is precisely the K-SAT problem formulated above.
Reduction to the K-SAT problem is a transparent way of representing the idea that multiple constraints need to be satisfied. The number K defines the gene redundancy and the probability of gene pleiotropy. Remind that pleiotropy occurs when one gene influences two or more seemingly unrelated phenotypic traits. The threshold h j and K define the number of genes which need be flipped in order to attain a high expression of the trait f j . Note that gene pleiotropy is a fundamental characteristics 37 , which is studied for real organisms only recently (see 19). We can compare experimental observations and consequence of model ( 2.4), which is a generalisation of K-SAT (compare plot Figure 1 and plots on Figure 1 in 19). So, we can fit our model parameters using real data. Moreover, we can check validity of our model by the following arguments.
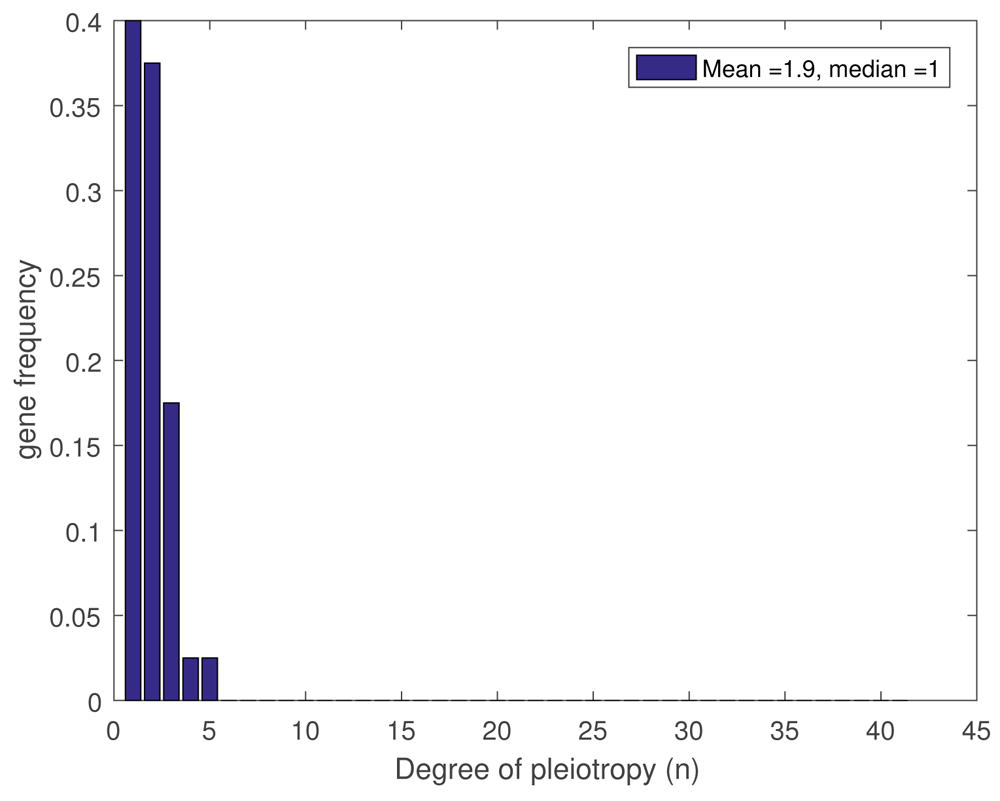
We note that, in the case of giant cluster formation, the topological properties of the solution set K , mentioned above, outline the properties of really observed fitness landscapes 22 : existence of many peaks, valleys and ridges connecting peaks. Namely, existence of many solutions of K-SAT, when a giant cluster exists, means that the landscape has a number of peaks separated by valleys. On the other hand, connectance of solutions within the giant cluster can be interpreted that there exist ridges that connect peaks.
Note that there are important differences between K-SAT in Theoretical Computer Science and fitness maximization problems. First, the signs of b j are unknown for real biological situations since the fitness landscape is unknown. Second, our adaptation problem involves the threshold parameters h j (see ( 2.4)). In contrast to K-SAT, in our case the Boolean circuit is plastic, because the h j are not fixed.
If the b j are unknown, the adaptation (fitness maximization) problem becomes even harder because we do not know the function to optimize. Therefore, many algorithms for K-SAT are useless for biological adaptation problems. Below we will nonetheless obtain some analytical results based on the assumption that b j are random.
The subsequent material is organized as follows. First we formulate a result on regulation mechanism power. Furthermore, we prove two fitness landscape learning theorems.
For simplicity, we consider asexual reproduction. To obtain similar results for sexual reproduction, one can consider a weak selection regime and use the results of 10, where eq. ( 2.9) are derived.
Let us introduce two sets of indices I + and I − , such that I + ∪ I − = {1,..., N b }. We refer to these sets in the sequel as positive and negative sets, respectively. We have
The biological interpretation of that definition is transparent: the expression of the traits f j with j ∊ I + increases the fitness and for j ∊ I – expression of the trait decreases the fitness.
Let s and be two genotypes. Then we denote by Diff( s, ) the set of positions i such that s i ≠ i :
The set Diff( s, ) indicates which genes in s should be flipped in order to obtain .
We formulate two theorems on fitness landscape learning. First we consider the case of infinitely large populations.
Theorem 3.1. Suppose that the evolution of the genotype frequencies X( s, t) is determined by equations ( 2.9) and ( 2.10). Moreover, assume that
I for all t ∊ [ T 1, T 1 + T c ] , where T 1, T c > 0 are integers, the population contains two genotypes s and such that the frequencies X( s, t) and X( , t) satisfy
II we have
for some j. In other words, the genes s i such that s i ≠ i are involved in a single regulation set K j ; and finally,
III Let
and
Then, if
we have j ∊ I +. If f j ( s) > f j ( ), then j ∊ I – .
Before proving this, let us make some comments. The biological meaning of the theorem is simple: for simple fitness models, where unknown parameters b j are involved in a linear way, in the limit of infinitely large populations fitness landscape learning is possible.
Moreover, note that we do not make any specific assumptions about the nature of mutation, but only that all genetic variation between s and are contained in a single regulatory set K j .
The assertion of Theorem 3.1 is not valid if the set Diff( s, ) belongs to a union of different regulation sets K j , j = j 1, . . . , j p with p > 1. This effect of belonging to different sets K j is pleiotropy in gene regulation. Note that if N b ≪ N g then the pleiotropy probability is small for large genome lengths N g . On the contrary, if N b ≫ N g then assumption II is invalid.
Assumption II looks natural if when we deal with point mutations. In fact, if is obtained from s by a single point mutation then condition ( 3.4) always holds for some j. For small mutation rates the probability of two point mutations is essentially below than the probability of a single mutation.
To conclude let us note that Theorem gives a rough estimate for the learning time T c :
Proof. The main idea is simple. Negative mutations lead to elimination of mutant genotypes from the population, and the corresponding frequencies become, for large times, exponentially small.
Assume that ( 3.7) holds. Let j ∊ I − , and thus b j < 0. Consider the quantity
According to assumption II
Assumption III entails that
Relations ( 2.6) and ( 3.10) imply
By ( 2.9) and the last inequality we find that for T > T 1
Consider inequality ( 3.11) for T = T 1 + T c . Let us note that in the relation Q( T 1) = X( s, T 1) /X( , T 1) the numerator is p 0 whereas the denominator ≤ 1. Thus, Q( T 1) ≥ p 0. The same arguments show that Q( T 1 + T c ) ≤ 1 /p 1. Therefore, by ( 3.11) one obtains that
This inequality leads to a contradiction for T c satisfying ( 3.6), thus completing the proof.
Theorem 3.1 can be extended to the case of finite populations and non-zero mutation rates. is small. To formulate this generalization, we need an additional assumption about the fitness function. Suppose that
where c F , C F > 0 are constants independent of t. For example, if
then c F = K F exp(– γ) and C F = K F exp( γ) and ( 3.13) holds if K F > exp( γ).
Condition ( 3.13) means that each individual gives birth to at least c F and at most C F descendants, where those bounds do not depend on the population size and evolution step.
Let
Note that for simplicity in the next Theorem 3.2 we consider point mutations (bit flipping) only. The model used here cannot represent mutations of arbitrarily small effect, but it can include insertions or deletions. In contrast to Theorem 3.2, Theorem 3.1 is valid for all kinds of mutations.
Then we have
Theorem 3.2. Consider the population dynamics defined by model 1 - 3 in Subsection 2.4. Assume conditions ( 3.14) and M hold, and assumptions ( 3.3), ( 3.4), ( 3.5), ( 3.7) of Theorem 3.1 are satisfied. Suppose
Then if j ∊ I – the inequality
is fulfilled with the probability Pr v such that
where for large N popmax and p mut → 0
Interpretation of Theorem 3.2
It is interesting to compare Theorem3.1 and Theorem3.2. The previous one asserts that for infinite populations the probability of the event j ∊ I − is zero whereas the second one claims that this probability becomes exponentially small as the population size increases.
This theorem also shows that evolution can make a statistical test checking the hypothesis H – that j ∊ I – against the hypothesis H + that j ∊ I +. Suppose that H – is true. Let V be the event that the frequency X( , t) of the genotype in the population is larger than p 1 within a sufficiently large time T c . According to estimate ( 3.18), the probability of the event V is so small that it is almost unbelievable. Therefore, the hypothesis H – should be rejected. We will refer T c as the checking time.
Ideas for the proof. The main idea is the same as that for the previous theorem: we compare the frequencies of the organisms with the genotype and the organisms with the genotype s. However, the proof includes a number of technical details connected with estimates of mutation effects and fluctuations. The formal proof can be found in Section 4. It is based on estimates of the accuracy of the Nagylaki equations (in the case of asexual reproduction).
Let us prove Theorem 3.2.
Let us introduce notation and make some preliminary remarks. Remind that we denote by N ( s, t) the number of the population members with the genotype s at the moment t. Let X( t) be the set of all population members at the moment t. For each x ∊ X( t) let us denote by ( x, t) the number of progeny born by the individual x at the moment t before the massacre (see point 3 of model from Subsection 2.4). Let s g ( x) be the genotype of x. Then, according to ( 2.8), the mean of ( x, t) is
where EX denotes the expected value of X. By ( s, t) we denote the number of all progeny born by individuals with the genotype s at the moment t before the massacre. Since all progeny are produced independently and randomly, the previous relation gives
Our main analytical tools are the Chernoff bounds and the Hoeffding inequalities. We also use the Markov inequality: for a positive random quantity X and a > 0 one has
Moreover, we use two elementary estimates. Let be an event in stochastic population dynamics. We denote by Not the negation (complement) of and by Pr( |ℬ) the conditional probability of under the condition ℬ. For events , ℬ 1, . . . , ℬ n we have
For two events , ℬ one has
Lemma 4.1. Let X i be independent random quantities, where i = 1, . . . , n. Let each X i be distributed according to the Poisson law with the average EX i = μ i. Let us denote
Then for all δ > 0
where
Similarly,
Proof. Note that for any λ > 0
Since X j are independent quantities, we have
The straight forward computation shows that
Therefore, due to the Markov inequality ( 4.3) and estimate ( 4.8) one has
where
We minimize f with respect to λ and obtain ( 4.6). To derive ( 4.7), we use
and repeat the same arguments. The Lemma is proved.
Lemma 4.2. Let X i be independent random quantities, where i = 1, . . . , n such that X i ∊ {0, 1} and EX i = p. Then
where
Proof. Note that for any λ > 0
Since X j are independent quantities, we have
Note that E exp( −λX j ) = p exp( −λ) + 1 − p. Let
We take λ = ln 2 and find that G(ln 2, p) = −g( p). Now by using the Markov inequality ( 4.3) and estimate ( 4.12) one obtains ( 4.10). The Lemma is proved.
We also use the following Chernoff-Hoeffding theorem. Let X i be i.i.d. quantities such that X i ∊ {0, 1} and EX i = p, where i = 1, . . . , n. Then for one has
where D( x||y) is the Kullback-Leibler divergence
Moreover, we will use the Hoeffding Theorem: if i.i.d. quantities X i ∊ [0, 1] with the probability 1 then
First we estimate the population size fluctuations.
Lemma 4.3. Let ( t) be the number of all progeny, born in the population at the moment t before the massacre, and ε 1 > 0 be a small number. Then
with probability
where
Proof. Let ( x, t) denote the number of progeny produced by the individual x before the massacre at the t-th evolution step. The number ( t) is the sum
of the mutually independent random quantities. According to ( 4.2), the average E ( x, t) is F( s g ( x)). Therefore,
We set
and use the Lemma 4.1 that gives us ( 4.17).
Lemma 4.4. Let ε 2 ∊ (0, 1) be fixed and condition ( 3.13) be fulfilled. Assume, moreover, that
where
and c F > 1 is defined by ( 3.13). Let us define the event ε 2 ( t) by
Then one has
where
and
Proof. Let ξ( x) be random quantities defined as follows: ξ( x) = 1 if the individual x is survived as a result of massacre (see point 3 of our model from Subsection 2.4), and ξ( x) = 0 otherwise. Let ( t) be the set of progeny produced by all individuals from the population. Then the number N sur( t) = N pop( t + 1) of finally survived progeny can be computed as follows:
Note that | ( t) | = ( t). Moreover, Eξ( x ) = N popmax/ ( t) for ( t) ≥ N popmax. Therefore, if ( t) ≥ N popmax then
Let us define the event
where the interval J ε ( t) is defined by ( 4.16) and is defined by ( 4.27). By ( 4.4) we have
Now we apply the Hoeffding inequality ( 4.15). For each ε 2 > 0 we obtain
If ℬ ( t) takes place, then ( t) ≥ N popmax and consequently
Therefore,
Moreover, by Lemma 4.3
Inequalities ( 4.30), ( 4.32) and ( 4.33) prove ( 4.25).
The following lemma, in particular, allows us to obtain equations ( 2.9) and ( 2.10) in the limit of infinite populations and for small mutation probabilities.
Recall that denotes the number of non-mutated progeny generated by the individuals with the genotype s before the massacre. Let N sur( s, t) be the number of those progeny that survived after that massacre.
Lemma 4.5. Let ε 0 be a positive number satisfying ( 4.75) and
Then one has
with the probability Pr s, t,+ such that
where
Similarly,
with the probability Pr s, t,− such that
Proof. Step 1, estimates of fluctuations. First let us estimate the fluctuations of the number . For each ε 2 > 0 let us define the event
By Lemma 4.1 one has
Note that
As a result, by ( 4.46) we obtain
Step 2. Here we estimate the number of progeny that survived as a result of the massacre procedure (point 3 of the population dynamics model, see subsection 2.9). Let X' ( s, t) be the set of progeny produced by individuals with the genotype s. Then the number N sur( s, t) of survived progeny x for individuals x belonging to the set Z' ( s, t) is
where ξ( x) are defined in the proof of the previous Lemma. For ε 3 > 0 we consider the event
Let us estimate the probability Pr( sur, s ( t)). According to the Hoeffding Theorem ( 4.15)
Note that ξ( x) and ξ( y) are independent quantities for different x and y, thus under the condition > N popmax
therefore,
Let us define the events ℬ s ( t) and ℬ( t) by
Then using ( 4.4) one has
We observe that under conditions ℬ s ( t) and ℬ( t)
In that estimate let us set ε 2 = 0.5 and ε 1 = 1. Taking into account that E = N pop( t) < 2C FN popmax, we have that
where
Moreover, according to ( 4.47)
and due to ( 4.17)
where R 1, R 2 are defined by ( 4.37) and ( 4.38). Finally,
Step 3, estimate of the number of mutants.
Let us estimate how many individuals with genotypes s can mutate. The probability of mutation is p mut. Let N mut( s, t) be the number of such mutants. Let us define the event mut, s ( t) by
Since the random quantity N mut( s, t) is subject to the Bernoulli law, we can apply the Chernoff-Hoeffding inequality ( 4.13). Then we obtain that
where, according to definition ( 4.14) of D( x|| y), one has
and g is defined by ( 4.11).
Using ( 4.4) one has
As a result, by Lemma 4.3 one finds
where R 4, R 5 are defined by ( 4.40) and ( 4.41).
To prove ( 4.35), we set ε 3 = ε 0/2. Taking into account condition ( 4.75) for ε 0 we see that if the both events Not mut, s ( t) and Not sur, s, ε 0/2( t) take place, then inequality ( 4.35) is fulfilled. Thus
where R i are defined by ( 4.37)–( 4.41).
Finally, taking into account the results of steps 1, 2 and 3 we see that estimate ( 4.35) holds with the probability Pr t,+. It completes the proof of ( 4.35). The second inequality ( 4.42) can be obtained in the same way.
We use the same idea that in the proof of Theorem 3.1 but first we establish uniform bounds for the population size and other quantities involved in the proof.
Step 1 Here we estimate the population size. Let us set
in Lemma 4.4. Let us consider the events ε 2 ( t) defined by ( 4.24) in Lemma 4.4. If the events ε 2 ( t) take place for all t ∈ [ T 1, T 1 + T c ] and N pop(0) = N popmax, we have that
Then conditions ( 3.15), ( 3.16) of Theorem 3.2 imply
Those inequalities imply the following estimates for the quantities R i defined by ( 4.37) –( 4.41):
where q i are defined by
where
and
where = 1 – ( κc F ) –1, and
For each p ∈ (0, 1) let us define an auxiliary function
where q i , are defined by relations ( 4.68)–( 4.72). We can find asymptotics of ρ under natural assumptions that p mut → 0 and N popmax → ∞ while all the rest parameters are fixed. Then the leading term in the right hand side of ( 4.36) is q 4 and As a result, we have
Step 2. Let Q( t) is defined by ( 3.8) and, moreover, let j ∈ I −. We use Lemma 4.5 inductively for genotypes s and . Let us set
where ε is defined by ( 4.75). We remark that the inequality
holds with a probability Pr Q, t > 0. Let us obtain a uniform estimate of that probability. Let ℰ( t) be the event that ( 4.78) holds at the step t. Using ( 4.4) we have
where, according to Lemma 4.4, the probability of the event Not ε 2 ( t) is less than η, where η is defined by ( 4.25), and
We conclude by ( 4.5) that
where is defined by ( 4.74). This estimate is uniform in t ∈ [1, . . . , T c ]. By ( 4.80) we obtain then that the inequality
is satisfied with the probability Pr v such that
For ε 0 defined by ( 4.75). one has
Now repeating the same arguments that in the end of the proof of Theorem 3.1, and taking into account asymptotics ( 4.77), we obtain the conclusion of Theorem 3.2.
In this paper, we proposed a model for fitness landscape learning, which extends earlier work by 7– 9 in two ways. First, we use hybrid circuits involving two kinds of variables. The first class of variables are real valued in the interval (0, 1) and can be interpreted as relative levels of phenotypic traits, other variables are Boolean and can be interpreted as genes. Second, we use a threshold scheme of regulation, which is inspired by ideas of the paper by 20. All variables are involved in gene regulation via thresholds.
The work presented here is a major extension of a long term effort to explicitly model the effects of phenotypic buffering in evolution by considering a class of Boolean and mixed Boolean-continuous models in which the phenotype is represented explicitly and the degree of phenotypic buffering can be controlled in various ways. For example, we have demonstrated that the idea of an “evolutionary capacitor” 38, 39 can be implemented by explicit control of phenotypic buffering in a hub-and-spokes architecture 23 and that in a more general class of genetic architecture numerical simulations show that an intermediate level of buffering is optimal for evolution in a changing environment 40 .
The results reported here are very promising, since they are consistent with the results of recent experiments by 41 and 42 on heat shock stress. The essential mechanism is that the exploration of the fitness landscape by the genetic network in such a way that future mutations are more likely to be adaptive. We have shown that, at least for some fitness landscapes, rapid evolutionary changes—perhaps instances of the “hopeful monsters” of Goldschmidt 43 —can be created by a combination of random small mutations and epigenetic effects. The main idea is that small mutation pave the way for large epigenetic or genetic changes. The hypothetical mechanism, which we propose, can be outlined as follows (see Figure 2, Figure 3).
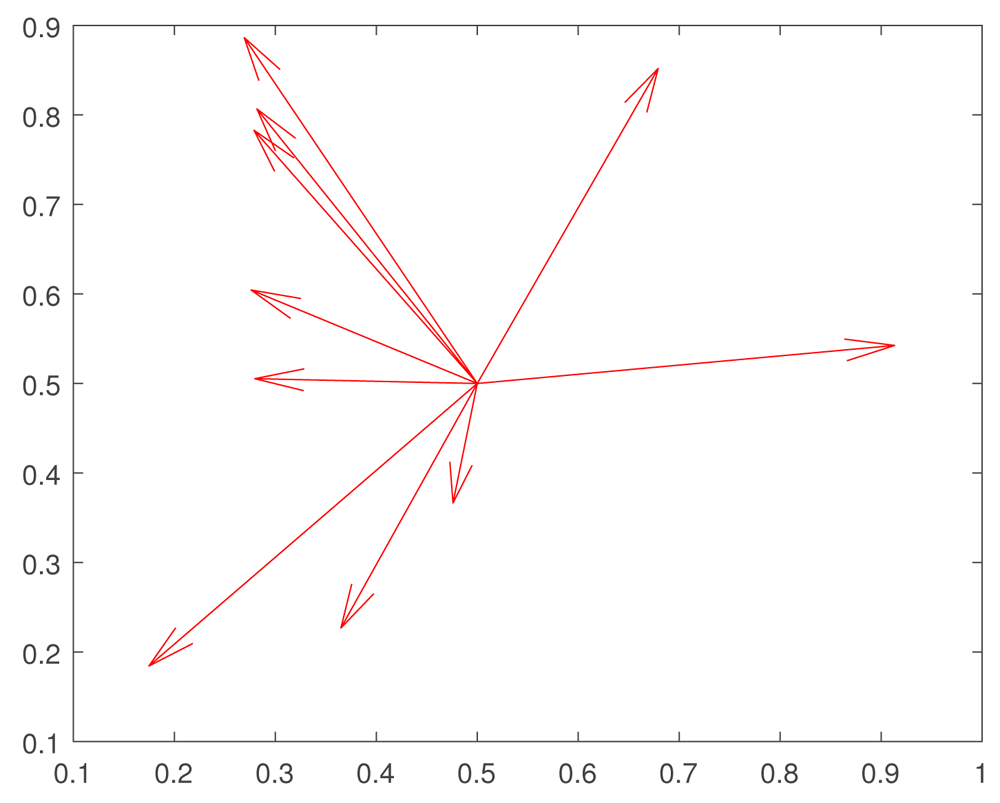
At initial moment the trait expressions take the values x = 0.5, y = 0.5. According to Fisher’s ideas, random large mutations decrease the fitness F = K F exp( W). (Changes of x = F 1, y = f 2, which are induced by mutations, are shown by red vectors.) Thus such mutations produce non-viable organisms.
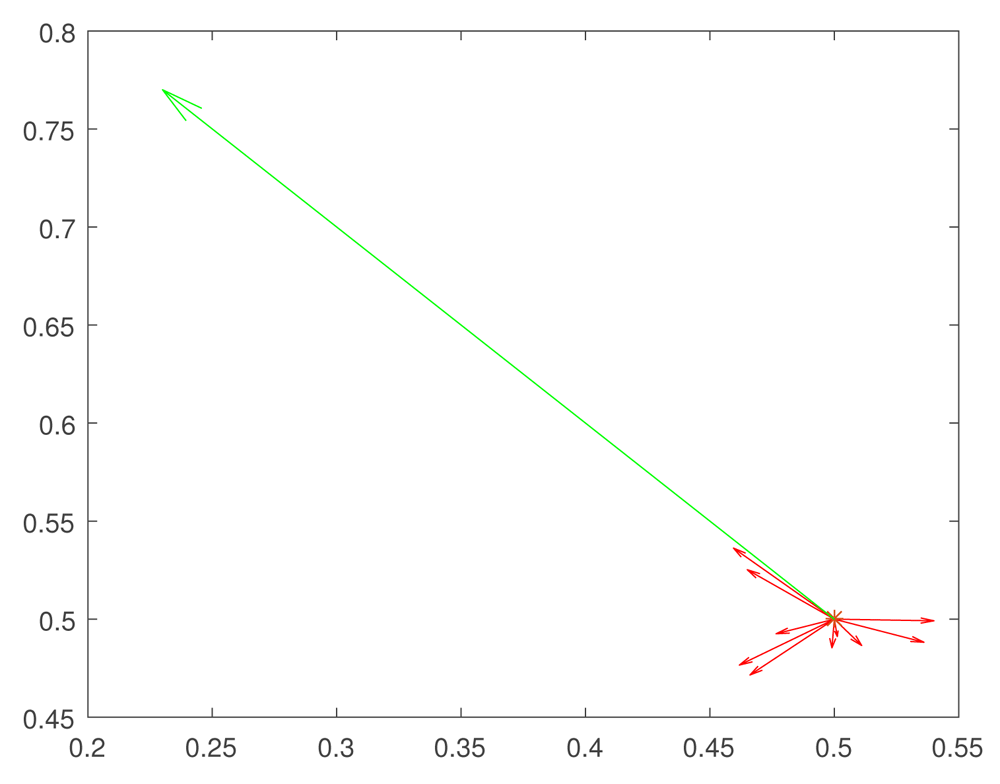
At the initial time the trait expressions take the values x = 0.5, y = 0.5. Evolutionary changes go in two stages. First we make small random mutations (shown by red vectors), which explore the fitness landscape. If such a mutation is not eliminated from the population, this means that a correct evolution direction is found, and gene regulation system makes a big leap (shown by the green vector) in the direction of that small mutation. Such a two step model can be called clever Goldshmidt leaps. Note that evolution is gradual, and the existence of clusters of almost identical genes involved in the same QTL increases the chances to create a clever Goldschmidt hopeful monster.
The expression of genes involved in the expression of phenotypic traits depends on threshold parameters h j , which take three values: a large negative one, a neutral value close to zero and a large positive one. First the threshold parameter h j is small and thus the phenotypic trait is sensitive with respect to even small mutations. Those mutations play a fundamental role working as scouts exploring environments (see Figure 3). If a mutation occurred and the corresponding mutant has survived within T c ≫ 1 generations then according to Theorem 3.1 and Theorem 3.2 these events mean that that mutation increases the fitness that allows the network to estimate the correct direction of evolution. Then gene regulation detects that increase to change the threshold according to simple rules. Namely, if the trait is less expressed in that mutant with respect to wild type parent, the gene regulation system decreases the threshold up to the large negative value. On the contrary, if the trait is strongly expressed in the mutant, the gene regulation system increases the threshold up to the large positive value. This simple regulation control not only sharply reduces the number of mutations needed for adaptation, but also canalizes the phenotype since for large thresholds the trait expression level becomes insensitive with respect to mutations. We suppose that these threshold modifications can be inherited.
So, we propose the mechanism: small mutations serve as scouts finding the way for large epigenetic or genetic changes, which can be performed by gene regulatory system.
The mechanism may also explain the results of 4 on prediction of environmental changes. In fact, let us suppose that environment varies in time. The first, perhaps relatively small, variations can trigger the threshold mechanism described above. As a result, the population will be adapted to the subsequent changes in advance.
Our results show that evolution can proceed rapidly because it reduces the number of mutations required for adaptive change.
The primary limitation of our results is that the representation of the evolving genetic network is limited to the network of gene controlling phenotype, represented here by the Boolean strings s. Other model variables represent the coarse-grained activities of genes. One class is the terminal differentiation genes represented by w ij , and another are the genes or epigenetic factors controlling the thresholds h and their associated learning rules. A more careful consideration of the relationship of these moieties to observable molecular entities is an important objective of future work. At the mathematical level, the key analytical results were obtained in a simplified context that falls short of a realistic level of pleiotropy and thus of the level of NP-hard complexity exhibited by fully pleotropic forms of our model. We believe that our analytical results can be generalized, which we plan to address in future work.
All data underlying the results are available as part of the article and no additional source data are required.
The second author was supported by the grant of Russian Ministry of Education, 2012-1.2.1-12-000-1013-016. Additionally, the second author was financially supported by Government of Russian Federation, Grant Grant 08-08. D. Grigoriev is grateful to the grant RSF 16-11-10075 and to both MCCME and MPI f\"ur Mathematik for wonderful working conditions and inspiring atmosphere. J. Reinitz and S. Vakulenko were supported by US NIH grant RO1 OD010936 (formerly RO1 RR07801).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
No source data required
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Hinton GE, Nowlan SJ: How Learning Can Guide Evolution. Complex Systems. 1987; 1.Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Theoretical/Mathematical Systems and Evolutionary Biology.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
No source data required
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Jiang P, Kreitman M, Reinitz J: The relationship between robustness and evolution. bioRxiv. 2018. Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Theoretical evolutionary biology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 13 Sep 19 |
read | read |
|
Version 1 01 Apr 19 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)