Keywords
CCAAT-box, distal promoter, NPR1 gene, RAV1AAT, W-box
This article is included in the Agriculture, Food and Nutrition gateway.
CCAAT-box, distal promoter, NPR1 gene, RAV1AAT, W-box
Some changes in our 2nd version dealt with the comments and suggestions addressed by the two reviewers Minghui Lu, and Santy Peraza-Echeverria.
In the Abstract: two bioinformatic tools: PlantCare and PlantPAN are added. The term “homology” has been changed to “identity”.
In the Methods: The “Genomic DNA isolation” subsection has been expanded. PlantCARE (Lescot et al., 2002) and PlantPAN are mentioned in the “Bioinformatic tools and sequence analysis” subsection.
In the Results and Discussion: Figure 1 has been replaced and now shows the transcription start site and the length of core promoter.
In the “Characteristics of the NPR1 distal promoter” subsection, we add a paragraph explaining that PCR based cloning strategy in combination with primer walking was applied to isolate the complete full length of putative distal promoter NPR1 gene region (5,950 bp). The terminology “99% homology” was changed to “99% identity”. We add that the BLAST analysis didn’t show any significant homology with another promotor sequence available in the NCBI database, indicating a limitation of promoter sequence availability. We further expand on this and add a reference to Lee et al., 2006.
We expand on our validation process and add a new reference (Nova et al., 2019).
We have updated Figure 3 with 1 TCA motif and 3 CGTA motifs in the cis-acting diagram and the word “nine” is changed to “eleven”.
We added a new paragraph explaining the findings following analysis with PlantCare and PlantPAN 3.0.
In the Conclusion: we add TCA and CGTA to the list of cis-acting elements and mention that the role of cis-acting elements in gene expressions needs to be confirmed.
See the authors' detailed response to the review by Santy Peraza-Echeverria
See the authors' detailed response to the review by Minghui Lu
The non-expressor of pathogenesis related gene 1 (NPR1) protein is the main regulator in the systemic acquired resistance response of many plants. Overexpression by modifying the distal promoter in the W-box element of the OsNPR1 gene in rice could increase its resistance against Xanthomonas oryzae pv. Oryzae up to 4.3 times (Hwang & Hwang, 2010). Similar results were reported by Zhong et al. (2015), who modified the RAV1 element on the GhNPR1 which increased Gladiolus hybridus resistance against Curvularia gladioli by up to 18.6 times compared to control. Based on those studies, we expect more prospects of the NPR1 gene promoter in the improvement of plant resistance against many pathogens.
Here we report characteristics of the distal promoter segment of the NPR1 gene isolated from chili pepper (Capsicum annuum) genotype Berangkai, a local genotype potentially produces more yields compared to other genotypes cultivated in West Sumatera.
Healthy young leaves collected from chili pepper genotype Berangkai was used as the plant material in this study. This genotype is known to produce more yield, but susceptible to geminivirus infection. Resistant genotype however, is not available in our collection so far, so comparison of both two genotypes is not possible to be performed. The plant was grown in a greenhouse and maintained for 8 weeks before being used for DNA isolation. Genomic DNA isolation was performed using protocol as described by Jamsari & Pedri (2013).
PCR-based cloning combined with a primer walking approach was applied for the isolation complete segment of distal promoter of the NPR1 gene. All PCR reactions in this study were performed using the KOD-Plus-Neo kit provided by Toyobo-Japan. Chromosome 7 of Capsicum annuum cv. Zunla-1 (Qin et al., 2014), accession number: NC_029983.1, was used as the reference sequence. Primer combinations were designed to cover the interval segment spanning from base 112.600.897 to 112.606.847 on the reference sequence (Table 1). All primers used in this study are listed in Table 1. First-step isolation was started from both termini and continued with successive isolation until both termini formed a complete contig of the target segment. The initial PCR condition was started with 95°C for 3 minutes for pre-denaturation and amplified in two different loops using 14 and 24 cycles. The first 14 cycles were performed using a touch down steps started by denaturation at 95°C for 30 seconds. Annealing was initiated with 70°C for 30 seconds and gradually decreased 1°C for every cycle before finally elongated in 72°C for 2 minutes. The next 24 cycles started with denaturation at 95°C for 30 seconds with annealing at 55°C for 30 seconds and elongation of at 70°C for 2 minutes and elongated for 72°C for 2 minutes. The final extension was maintained at 72°C for 5 minutes. PCR reaction was performed in 50 µl of final volume. PCR composition was set according to manufacturer’s recommendation, containing 5 µl 10x Buffer KOD-Plus-Neo, 5 µl of 2 mM dNTPs, 3 µl of 25 mM MgSO4, 1.5 µl of 10 ng/µl of each primer, 1 µl of KOD-Plus-Neo (Toyobo, Japan) and 1 µl of 10 ng/µl of DNA template. The volume was filled with 32 µl of nuclease-free water.
The PCR product generated from each walking step was processed to sequencing reaction using its both forward and reverse primers. All sequencing processes were performed by 1st BASE-Singapore. A number of bioinformatic tools were applied during analysis of the sequence. Trimming, editing and building of the sequence contig were performed using BioEdit v7.2.5 (Hall, 1999). Homology search of the sequence with all available sequences deposited in the NCBI database was run using the BLASTn tool (Altschul et al., 1990). The cis-acting elements were identified using PLACE, developed by Higo et al. (1999), PlantCARE (Lescot et al., 2002) and PlantPAN.
We successfully isolated the distal promoter of the NPR1 gene from our local chili pepper genotype Berangkai, exhibiting 5,950 bp in size and designated PD_CbNPR1. The fragment was isolated from two steps round of walking via PCR-based cloning. In the first step, the whole distal promoter region was expected to be covered using two primer combinations (F1 F/R and F2 F/R). However, after verification of the sequence data using BioEdit, the upstream primer combination (F1 F/R) successfully produced a contig spanning only 1,963 bp, while the downstream primer combination (F2 F/R) was skipped for the sequencing process. In the second round of primer walking, four new primer combinations (F1.1-F/R, F1.2-F/R, F2.1-F/R, and F2.2-F/R) (Table 1) were designed in order to extend sequence coverage. Combining all verified sequences obtained from the first and the second round of primer walking successfully produced a contig with a size of 5,950 bp (Figure 1).
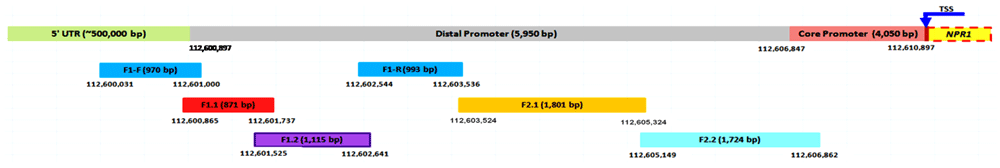
Base position of start and end segment are shown by number on each terminus. Length of verified sequence of each segment is shown by number in bracket on each segment. Transcription start site of the NPR gene located between core promoter and NPR1’s ORF is shown by arrowhead.
PCR based cloning strategy in combination with primer walking was applied to isolate the complete full length of putative distal promoter NPR1 gene region (5,950 bp). This approach is considered to be the most appropriate since the sequencing read capacity used in this study is limited for about only 500 bp on average. Even though the KOD-Plus-Neo could amplify up to 24 kb according to manufacturer’s claim, but the full-length fragment (5,950 bp) still can not be sequenced in one step read due to limited reading capacity of the sequencing machine. Validation of every single nucleotide data was confirmed by at least two overlapping validated segment.
Homology analysis between PD_CbNPR1 and its reference sequence via BLAST search showed 99% identity. The data was verified by only one single nucleotide polymorphism (SNP) shown as a substitution event at the position -6,335 from the ATG start codon (Figure 2). Furthermore, BLAST analysis showed no significant homology with another promotor sequence available in the NCBI database, indicating a limitation of promoter sequence availability. The only promoter sequence showed homology is the promoter region of Capsicum annuum pathogenesis related protein-1 (PR-1) gene (DQ201633.1) published by Lee et al. (2006). However, the comparable nucleotide of both sequences spanned only 180 bp.

In order to validate our claim, we constructed a contiguous segment with our isolated core promoter (MK310185.1) and the NPR1 gene sequence isolated from a similar genotype Berangkai (Nova et al., 2019). BLAST analysis using the NPR1-Berangkai cDNA sequence exhibited 43 significant identity hits with other cDNA sequences of NPR1 gene for instance, Capsicum annuum (NM_001325099.1), Capsicum chinense (AM900559.1), Solanum lycopersicum (KX198701.1, NM_001247629.2), Nicotiana sp. (DQ837218.1, AF480488.1) Carica papaya (XM_022041103.1, AY550242.1) and some others. Tree analysis showed that our NPR1-Berangkai cDNA sequence clustered to similar clade with AM900559.1 and NM_001325099.1 and other three solanaceae (S. lycopersicum-KX198701.1, NM_001247629.2; S. tuberosum-XM_006357647.2; S. pennellii-XM_015227358.2 and S. torvum-KJ995663.1). All those data obviously indicated that our segment landed in the right chromosome segment.
PLACE analysis successfully indicated 9 cis-acting elements with multi repetition (Figure 3). All 9 cis-acting elements contained the W-box (15), WLE1 (W-box like elements) (8), RAV1AAT (20), TATA-box (22), CAAT-box (26), GARE (1), GT1 (20), Enhancer (4) and Silencer (1). The PlantCare analysis successfully showed 2 other cis-acting element motifs namely 1 TCA motif and 3 CGTA motis which could not be shown by PLACE (Figure 3).
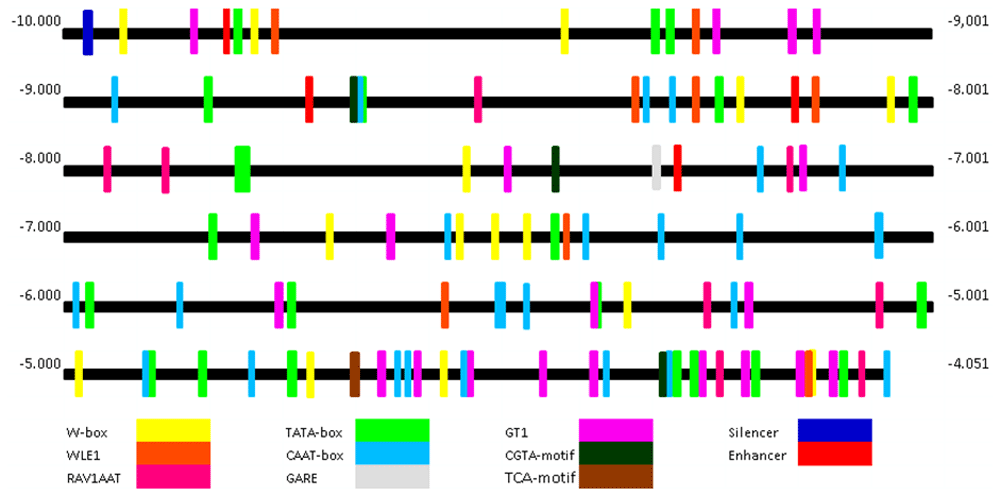
Colors indicate each element as labeled below the figure.
The W-Box element binds the WRKY protein, acting as a transcription factor during expression of NPR1 (Yu et al., 2010). Mutation in this element in Arabidopsis thaliana delayed the NPR1 expression induced by salicylic acid (SA). PLACE analysis indicated that PD-CbNPR1 is characterized by three consensus sequences: TTGAC, TGACY, and TGACT. The TGACY pattern could also be found in the promoter of ERF3 isolated from tobacco (Nishiuchi et al., 2004), while TGACT could be found in the promoter of the Iso1 barley gene (Sun et al., 2003). The WLE1 (W-box like elements) with TGACA pattern has an analog function with the W-box identified in the promoter of OsNPR1 (Hwang & Hwang, 2010).
Another cis-acting element found in PD_CbNPR1 is RAV1AAT, which acts as binding site for protein RAV1 (Hwang & Hwang, 2010). The RAV1 protein is a transcription factor associated with some pathogenesis-related genes (Sohn et al., 2006). Two consensus patterns of RAV1AAT elements found in PD_CbNPR1 are CAACA and TGTTG. The CAACA pattern was also found by Kagaya et al. (1999) in Arabidopsis thaliana, while the TGTTG pattern is similar to the OsNPR1 promoter (Hwang & Hwang, 2010). Notably, the ASF1 element is absent in the PD_CbNPR1. The ASF1 element has been previously reported by Hwang & Hwang (2010) and Zhong et al. (2015) as a cis-acting for common promoter associated with pathogen infection and SA induction.
PLACE analysis identified 22 putative TATA-boxes in our PD_CbNPR1, which could be classified into TATA box 3, TATA box 4, TATA box 5 and TATA box OspaI. During transcription, the TATA-box binds with RNA polymerase II and many transcription factors like TFIID, TFIIA, TFIIF, TFIIE, and TFIIH to build the TATA-box complex.
Another interesting element found in PD_CbNPR1 is the CAAT-box (26 repeats) which is associated with the regulation of many genes involved in pathogen infection (Imran et al., 2016). The CAAT-box consensus sequence found in PD_CbNPR1 is characterized with CAAT which is similar that have been found in promoter of the legA gene from Pisum sativum (Shirsat et al., 1989).
We also found a gibberellin-responsive element (GARE) motif, which has been previously reported by Ogawa et al. (2003) as the binding site with some transcription factors induced by gibberellic acid. The consensus sequence of thoses motifs is TAACAAR. Another cis-acting element induced by abiotic factors such as salt, light and pathogenesis is GT1 motif (Biłas et al., 2016; Park et al., 2004). The identified GT1 element in PD_CbNPR1 is characterized by a consensus GRWAAW motif and present in 20 repeats. They could be classified into two forms, GT1CONSENSUS and GT1GMSCAM4, as described previously by Terzaghi & Cashmore (1995). The last GT1 class, GT1GMSCAM4, is initially identified by Park et al. (2004) with consensus pattern GAAAAA.
PLACE analysis also successfully identified four CCAAT-box motifs, which is commonly reported as an enhancer of transcription (Thonpho et al., 2013). This element is able to up-regulate the transcription and replication rate of eukaryotic genes by binding many types of transcription factors (Biłas et al., 2016). da Silva et al. (2013) previously reported that a greater number of CCAAT-boxes could improve promoter capability in increasing of replication rate by binding efficiently with transcription factors. In contrast with the enhancer, we found only one motif of GAGAAATT which is also known as silencer (Lai et al. (2009). This element is reported to be able to down-regulate the kinesin-like protein 1 (AtKP1) gene in Arabidopsis thaliana by up to 80.9%.
Analysis with PlantCare (Lescot et al., 2002) resulting 2 new additional elements, which are designated TCA element and CGTA-motif (Figure 3), while analysis with PlantPAN 3.0 did not show any new different element compared to PLACE analysis. The TCA element has a function as a cis-acting element which is involved in salicylic acid responsiveness. Moreover we also found the CGTA motif playing a role in regulating me-JA responsiveness. In our sequence only 1 single TCA element and 3 CGTA motifs could be detected.
We successfully isolated the distal promoter of the NPR1 gene from chili pepper genotype Berangkai (designated PD_CbNPR1) spanning 5,950 bp. The PD_CbNPR1 contain 11 cis-acting elements: W-box, WLE1, RAV1AAT, TATA-box, CAAT-box, GARE, GT1, TCA, CGTA, enhancer and silencer elements. Engineering of the cis-acting elements may have future prospects, particularly in improving chili pepper resistance against biotic and abiotic stresses by up- and down-regulating NPR1 gene expression. However, their role in the gene expression has to be confirmed empirically. For that reason, we are currently using the cis-acting elements found in this study for replicase (Rep) gene expression in the bacterial system.
The sequence of the distal promoter of the NPR1 gene sequenced in this study has been deposited in GenBank under accession number MK281381; https://identifiers.org/ena.embl:MK281381.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Nova B, Hidayati R, Fichri M, Syukriani L, et al.: Isolation and characterization of the NPR1 gene from highly susceptible pepper cv. Berangkai.Bioscience Research. 2019; 16 (2): 1317-1322 Reference SourceCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Plant molecular biology and plant physiology
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: plant tolerance to abiotic stresses especially high temperature
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular Plant-Pathogen Interaction and Plant Biotechnology
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: plant tolerance to abiotic stresses especially high temperature
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 24 Sep 19 |
read | read | |
|
Version 1 14 Jan 19 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)