Keywords
morphogenesis, signaling pathways, membrane traffic, secretion, Spitzenkörper, host-interactions, cellular organization
morphogenesis, signaling pathways, membrane traffic, secretion, Spitzenkörper, host-interactions, cellular organization
Morphology changes occur in a range of human fungal pathogens upon interaction with the host1. In response to different host signals, Candida albicans switches from the yeast form to a hyphal form, a cell shape characteristic of filamentous fungi, such as Aspergillus nidulans and Neurospora crassa2–4. However, hyphal cells of C. albicans are different from those of these organisms with respect to shape/diameter and extension rate (10- to 100-fold slower with this fungal pathogen). Furthermore, in these filamentous fungi, microtubules are critical for hyphal growth, a striking difference with C. albicans, in which microtubules do not play a prominent role5. C. albicans is an opportunistic human fungal pathogen and a number of studies have linked the switch from yeast to hyphal form with pathogenicity, whether during superficial or systemic infections6–10. This brief review presents an update of research from the past 2 to 3 years on C. albicans technological advances, cell signaling, host interactions, and membrane traffic and puts an emphasis on hyphal growth (Figure 1).
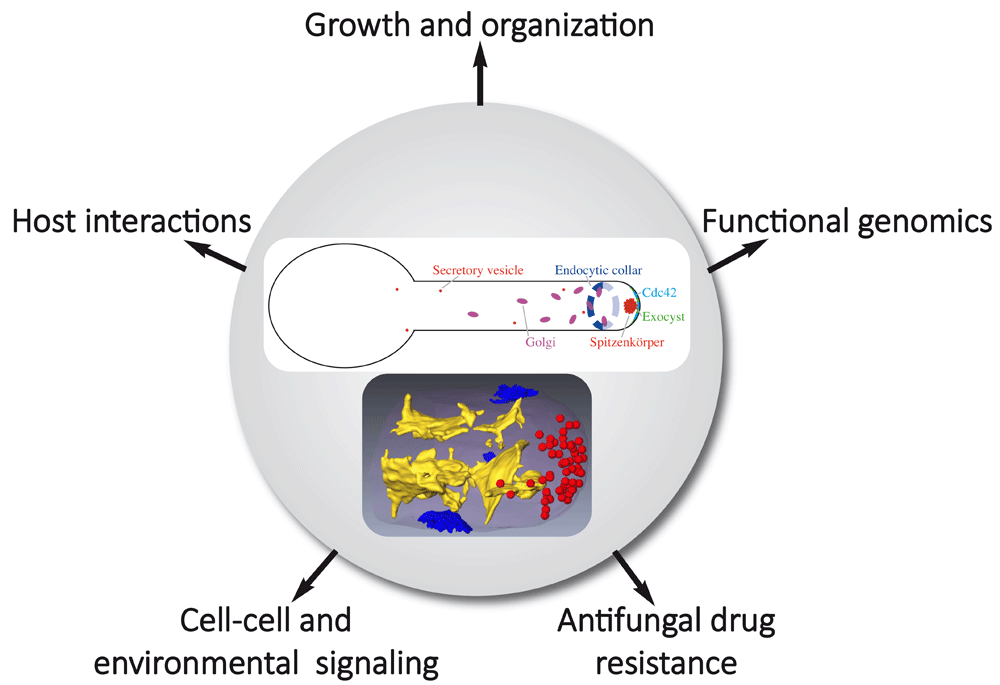
The upper panel shows membrane compartments of the exocytic and endocytic pathways focusing on compartments discussed in the review. Endoplasmic reticulum and endosomes, for example, are not shown. The lower panel, reproduced from Weiner et al.33, illustrates a segmented three-dimensional dataset from focused ion beam/scanning electron microscopy tomography of a hyphal tip with internal membranes (yellow), secretory vesicles (red), and sites of endocytosis (blue).
In the past several years, technological advances have opened a range of new possibilities in C. albicans research. Specifically, the majority of approaches have opened our horizons with respect to large-scale analyses of fungal pathogen function, including a major thrust coming from clustered regularly interspaced short palindromic repeat (CRISPR)-based tools that have particularly revolutionized genome manipulation in genetically less accessible fungi, such as the diploid C. albicans11,12. Other notable approaches that are changing how we work with and view this fungal pathogen include experimental or micro-evolution approaches13–15, in particular with respect to host niche environments. In addition, large-scale approaches, such as population and genetic diversity analyses via genome sequences of large numbers of isolates16, and the establishment of genomic platforms that facilitate the study of gene function at a genome-wide level17–20 pave the way for future multi-omic studies.
The application of CRISPR-based methods to C. albicans in 2015 was a major step in facilitating molecular genetics in this less genetically tractable fungus21 and opened a myriad of possibilities for studying gene function, including marker recycling22,23, a “gene drive array” platform for genetic interaction12, rapid gene concatenation for genetic rescue of multi-gene mutants24, and gene regulation25,26. Overall, C. albicans CRISPR-based methods have been substantially optimized11,12,23,25–28 and now facilitate a range of gene functional analyses up to a genome-wide scale.
Experimental or micro-evolution approaches are particularly powerful tools when applied to opportunistic pathogens29. These approaches have been used initially to identify mutations that restore filamentation in a non-filamentous mutant within macrophages30 and more recently to investigate drug resistance14, host niche-specific mutations13, and the emergence of mutualism between host and fungus15. These approaches, coupled with whole-genome sequencing and other genome-wide methods, are extremely useful when applied to a diploid commensal that can undergo a panoply of genome rearrangements with far-reaching consequences.
The application of novel large-scale approaches, as well as the refinement and optimization of existing methods to gene function analyses in C. albicans, will undoubtedly promote a deeper understanding of this fungal pathogen. Chemical inhibitors and chemogenomic profiling have been used to identify genes involved in enhanced antifungal drug sensitivity or resistance31 and novel inhibitors of morphogenesis32.
Genome sequencing and comparative genomics of 182 worldwide C. albicans isolates have revealed evidence of gene flow and a highly clonal lineage that has undergone substantial pseudogenization16. In 2018, three major studies highlighted advances made possible by new gene function platforms and tools17–19. Two groups took advantage of powerful transposon-based approaches, coupled with a stable haploid C. albicans derivative, to probe essential genes, generate a comprehensive set of mutants in this fungus, and carry out genome-wide screens17,19. These studies yielded important information on gene essentiality and azole resistance in this fungal pathogen. Extensive effort was also invested in generating a genomic platform centered on an ORFeome collection representing the majority of open reading frames (ORFs) in Gateway donor vectors, together with a wide range of expression vectors18 facilitating genome-wide overexpression analyses and protein–protein interaction studies16,20,34. Together, these new technologies have facilitated recent advances in hyphal growth signaling, host interactions, and membrane traffic.
In the past 2 to 3 years, a range of studies have investigated hyphal growth signaling in C. albicans35. These studies have made significant advances, in particular in the areas of amino acid inducers of the hyphal transition36,37, gaseous sensing and signaling38–42, and reactive oxygen and oxidative stress signaling43–45. Extensive analyses of filamentation programs revealed media-independent genetic requirements for filamentation, in particular RIM101 (pH-dependent pathway) and GPA2 (Gα functioning in the cAMP/PKA pathway), in addition to a core transcriptional profile46. Also, an investigation into the cAMP requirement for hyphal morphogenesis showed that basal levels of cAMP are sufficient for hyphal formation in response to N-acetylglucosamine (GlcNAc), suggesting that cAMP-independent signals are also important for hyphal induction47. Both G1 and S phase arrest can induce filamentous growth and this has been shown to require the cAMP/PKA pathway48.
Nutrient deprivation triggers hyphal development in C. albicans, and various amino acids have been shown to be critical for this transition. The groups of Van Dijck36 and Ljungdahl37 investigated cAMP/PKA-dependent morphogenesis that is triggered by arginine, ornithine, proline, and methionine metabolism. For these different amino acids, induced expression of amino acid permease genes is critical, with the former three amino acids being metabolized in the mitochondria, resulting in elevated ATP levels that appear to increase activation of the Ras1/cAMP/PKA pathway. With respect to methionine, it is converted to S-adenosyl methionine (SAM) that is subsequently decarboxylated and the resulting amino-propyl group is converted to polyamines that have been shown to activate adenylate cyclase.
Although a number of studies have previously analyzed the roles of oxygen and CO2 signaling in hyphal development, there has been little attention to nitric oxide (NO) signaling in this process. Koch et al. examined a metabolic checkpoint for the yeast-to-hypha transition that is regulated by endogenous NO signaling and their results indicate that sufficient endogenous NO releases Nrg1 repression of this transition38. Three recent studies have shed light on how the tricarboxylic acid (TCA) cycle regulates CO2 signaling42, how a phosphatase–kinase pair controls CO2-responsive Ume6 phosphorylation and stability39, and have identified a link between CO2 sensing and lipid/Pkh1/2 kinase signaling during hyphal development41. In the first of these studies, the authors used a library of TCA metabolic pathway mutants to show that the TCA cycle plays a critical role in regulating CO2 sensing and hyphal development42. Lu et al. carried out a genetic screen to determine the CO2 signaling pathway that regulated Ume6 stability and found that a kinase–phosphatase couple controlled the CO2 response of this transcription factor that is crucial for hyphal elongation39. A screen in Saccharomyces cerevisiae for mutants that regulate the transcription factor Cst6 (C. albicans homolog Rca1), which activates the carbonic anhydrase NCE103 in a CO2-dependent fashion, identified the kinase Sch941. The authors went on to show that Sch9 phosphorylates the transcription factor Rca1 in C. albicans and that it links CO2 adaptation to lipid signaling via Pkh1/2.
The production of reactive oxygen species (ROS) during C. albicans morphogenesis plays an important role in pathogenicity. The conserved heat shock factor-like transcriptional regulator Skn7 is critical for filamentous growth and protection from the accumulation of intracellular ROS in these conditions43. Interestingly, a member of the NADPH oxidase (NOX) family, Fre8, was recently shown to produce a ROS burst during morphogenesis, which is particularly important in the animal host45. Recent studies by Liu et al. have shown that inhibition of the major high-affinity phosphate importer, Pho84, sensitized C. albicans to oxidative stress via inducing ROS accumulation through activation of TOR (target of rapamycin) signaling44. In addition to these environmental conditions, quorum-sensing molecules, such as farnesol, regulate the morphological transition, and recent work proposed that the response of C. albicans to farnesol is influenced by Eed1, a protein critical for hyphal growth maintenance49. Together, these different advances in hyphal growth signaling highlight the important role of hyphal development in host niches and in response to a range of relevant host signals.
The microbiota is thought to, in part, restrict the fungus to the commensal state50. Of note, the GUT (gastrointestinally induced transition) cells, which are postulated to be a specialized commensal form in the mammalian gastrointestinal (GI) tract, are less virulent in a mouse bloodstream infection model51,52. The alteration of the balance between commensalism and pathogenicity in the presence of the gut microbiota is associated with mutations in C. albicans transcription factors required for white-opaque switching and filamentation, such as Efg1, Wor1, and Flo815,52,53. Furthermore, C. albicans strains that are hyperfit in the antibiotic-treated or germ-free mouse gut tend to be deficient in hyphal morphogenesis53,54, yet the observation that a hyperfit ume6 mutant has a ratio of yeast and hyphae similar to that of the wild-type strain in the mouse GI tract would argue that cell shape per se does not determine commensal fitness55. Using an experimental system based on long-term GI tract colonization of mice, a recent work nicely demonstrated that in the absence of microbiota C. albicans evolves into strains that lose their ability to form hyphae15. Interestingly, this study additionally shows that priming naïve mice with the gut-evolved strains resulted in a broad cross-protection against Aspergillus fumigatus, Staphylococcus aureus, or Pseudomonas aeruginosa.
Host defense also includes the epithelial physical barrier and host immune cells, such as macrophages. Hyphal growth is associated with mechanical forces during the interaction of C. albicans with such host cells56. The relative contribution of these mechanical forces to host cell damage, compared with other hyphal attributes, is an area of active investigation. Mechanical forces appear to be sufficient to penetrate epithelial cells even in the absence of secreted factors, such as the toxin candidalysin, encoded by ECE1 and therefore secreted only by hyphae57. Indeed, ece1Δ/Δ mutant hyphae can invade intestinal epithelial cells without causing damage, yet optimal damage induction requires a combination of hypha formation and candidalysin secretion58. The ability to undergo yeast-to-hypha morphogenesis and the cell wall composition are also important determinants in the macrophage–C. albicans interaction59. Recently, C. albicans escape from the phagolysosome was proposed to rely directly on physical rupture60. C. albicans cells induce macrophage cell death via pyroptosis, a caspase-1–dependent programmed cell death61,62, and it was proposed that activation of the inflammasome is a consequence of this phagolysosome rupture via the yeast-to-hypha transition60. However, this proposal was challenged by another study, which showed that rupture is not a prerequisite for inflammasome activation, as a collection of genes enabled activation of macrophage pyroptosis independently of effects on morphogenesis, and cell wall remodeling was a major determinant63. The role of candidalysin in the phagocyte inflammatory and damage response to C. albicans hyphae was recently investigated and this toxin appears to trigger inflammasome activation64. Thus, how C. albicans morphological transition, phagosomal neutralization and rupture, and pyroptosis are linked remains a topic of active research65,66.
Secretion plays an essential role during C. albicans virulence, in releasing candidalysin and a variety of proteases and lipases. In addition to using the conventional secretory pathway to secrete components into the external medium, similar to other fungi, C. albicans releases extracellular vesicles (EVs)67, which contain cytoplasmic and moonlighting proteins, and membrane and cell wall–related proteins68. A recent study elegantly showed that the EV population and composition released by C. albicans during growth in a biofilm are distinct from those of planktonic cells69. In particular, as exogenous delivery of wild-type vesicles restores the biofilm drug-resistant phenotype and matrix composition to a subset of ESCRT (endosomal sorting complexes required for transport) mutants, it was proposed that biofilm EVs, which consist predominantly of a 30- to 200-nm diameter population, corresponding in size to exosomes, have a direct role in matrix biogenesis and carry specific cargos to confer drug resistance. The mechanism by which EVs would reach the matrix is still unclear. However, a recent work shows that AmBisome (60 to 80 nm liposomes) can traverse the cell wall70, suggesting that EVs may also directly transit the cell wall.
Rapid hyphal growth requires active endocytosis to counterbalance exocytosis at the hyphal tip and recycle membrane lipids and proteins71,72. For example, recent work demonstrated that polarization of a chitin synthase to the hyphal apex in A. nidulans occurs by indirect endocytic recycling73. Genetic analyses of loss-of-function mutants in a number of genes implicated in actin regulatory complexes, such as Pan174 and Myo575–77, have also confirmed the importance of endocytosis in C. albicans hyphal growth, and two recent articles further point to an increased requirement for endocytosis during hyphal growth, compared with budding growth. Taking advantage of a complete collection of kinases and phosphatases, regulated via an inducible TETon promoter, Bar-Yosef et al.78 identified a novel regulator of hyphal morphogenesis, Akl1 (related to the Ark/Prk family of kinases79), whose overexpression reduced hyphal extension rates and conversely whose deletion resulted in an initial increase in hyphal extension rate. Furthermore, screening of well-characterized drug libraries allowed the identification of specific inhibitors of hyphal morphogenesis, related to piperazine32. Although these drugs inhibited hyphal formation at concentrations that appear to be above safe levels, these studies raise the prospect of identifying molecules that target fungal endocytosis as potential inhibitors of C. albicans virulence.
Membrane/protein trafficking to the plasma membrane is mediated by vesicular transport between different cellular compartments, and small GTPases of the Arf (ADP-ribosylation factor) and Rab (Ras-related in the brain) families regulate each step of these processes80–82. The role of Arf proteins was recently investigated in hyphal growth and virulence. Of the five Arf/Arl proteins, Arf2 and Arl1 were shown to be critical for virulence in murine models for candidiasis, and Arl1 was more specifically required for oropharyngeal candidiasis83. In addition, an arf1 mutant was shown to exhibit reduced virulence in a murine systemic infection model and in macrophage killing yet this strain had a reduced growth rate and underwent cell cycle arrest84. In the latter study, Arf1 was implicated as a regulator of endoplasmic reticulum (ER)–mitochondria interactions, which would directly or indirectly impact ERMES (ER–mitochondria encounter structure). Whereas Arf2 is required for viability, Arl1 is involved in hyphal extension and in restricting hyphal growth to a single site. The hyphal extension defect of the arl1 mutant was associated with an altered distribution of the Rab GTPase Sec4 and both defects could be restored by overexpression of the Rab GTPase Ypt683,85, suggesting that a genetic interaction between Arl1 and Ypt6, perhaps via the GARP (Golgi-associated retrograde protein) complex86, could be specifically critical for hyphal growth. In S. cerevisiae, analysis of trafficking mutants demonstrated that the late stage of exocytosis is particularly critical to regulate endocytosis87, and more recently it was shown that Sec4 coordinated polarized exocytosis with the assembly of cortical actin patches that initiate endocytosis88, indicating that this Rab GTPase is central for the balance in membrane trafficking.
Individual Rab GTPases can coordinate multiple transport pathways by recruiting effectors to different organelles89, and the importance of Rab GTPases during hyphal growth has been investigated in filamentous fungi, such as A. nidulans and N. crassa3,90. However, as mentioned above, the differences in hyphal growth in these fungi, compared with C. albicans, raise the question as to how hyphae are organized to regulate membrane traffic in this organism (Figure 1). Using three-dimensional electron microscopy, a high-resolution view of the C. albicans hyphal filament shows that the secretory pathway is organized in three distinct structural domains: sheet-like parallel membranes, shorter sheet-like membranes, and the Spitzenkörper (Spk), which is composed of a uniform population of approximately 60 vesicles that are about 70 nm in diameter33. Thus, the C. albicans Spk appears to be simpler than that of filamentous fungi, which is composed of a heterogeneous population of vesicles2–4. Dynamic analyses of vesicle delivery to the apex suggest that short-range vesicle delivery significantly contributes to filamentous growth in C. albicans and that the Spk could act as a focal point for incoming secretory vesicle traffic, produced in the subapical and apex regions33. These distinctions between the Spk of C. albicans and that of filamentous fungi might reflect differences in their function. In particular, a characteristic shape change of the Spk, from globular to crescent-like, appeared to be associated with increased extension rate in A. nidulans, as secretory vesicles accumulated at the Spk during phases of slow growth subsequently fused with the plasma membrane91. Such a stepwise growth mode in hyphae has been shown in several filamentous fungi92,93 but thus far not in C. albicans.
Overall, this broad range of findings in the past several years has provided both exciting novel approaches and new research directions that give us insight into the biology of this fascinating fungal pathogen. As we understand, in greater detail, the basic biology of this fungus, we now can put this new knowledge into the context of the host and the balance between commensalism and infection. Without a doubt, the advent of new technologies, in particular the combination of large-scale approaches, and effectively mixing and matching them with animal-based studies will provide powerful platforms for novel gene discovery and functional analyses in the years ahead.
This work was supported by the Centre National de la Recherche Scientifique, the Agence Nationale de la Recherche (ANR-16-CE13-0010-01 and ANR-11-LABX-0028-01), and the European Union H2020 (MSCA-ITN-2015-ETN-GA-675407) grants.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
|
Version 1 21 May 19 |
read | read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)