Keywords
Orofacial cleft, cleft lip with or without cleft palate, case-parent triads, gene-environment interaction, parent-of-origin, PoOxE, Haplin
Orofacial cleft, cleft lip with or without cleft palate, case-parent triads, gene-environment interaction, parent-of-origin, PoOxE, Haplin
In this version we have considered the remarks of Dr. Evie Stergiakouli. We have addressed some limitations that were pointed out and generally edited the manuscript for clarity.
Abstract:
Removed reference to previously published work and added information about the methodology.
Introduction:
Rephrased sentence that overemphasized how intriguing the problem of missing heritability is in clefting.
Methods:
Clarified that 59 individuals of other ethnicities than Asian or European were included in the pooled sample and added a sentence about the American Statistical Association’s negative attitudes towards using a fixed p-value as a threshold for statistical significance.
Results:
Revised titles of Tables 3, 4 and 7 to include information about which analyses were conducted.
Discussion:
Added a paragraph about genomic imprinting as a possible mechanism for PoO effects, clarified that our findings should be treated with caution until they have been replicated, and addressed the limitation that we have not imputed genotypes.
In general:
Corrected minor errors and slightly altered text to improve flow and precision.
See the authors' detailed response to the review by Evie Stergiakouli
Cleft lip with or without cleft palate (CL/P) appears in approximately 3.4 to 22.9 per 10,000 live births1. Based on the severity of the cleft, patients undergo varying degrees of medical, dental, speech and psychosocial interventions over the first two decades of their lives, a long-term multidisciplinary treatment that not only imposes a heavy burden on patients and their families2,3, but also accounts for a substantial outlay in national healthcare budgets4,5.
Multiple genetic and environmental factors have been reported to influence the risk of CL/P, individually and through complex interactions in relevant biological pathways6–10. Major advances in high-throughput genotyping technologies, coupled with a boost in international collaborations, have led to substantial progress in gene-mapping for orofacial clefts, and the first wave of genome-wide association studies (GWAS) identified and replicated several key genes and loci associated with clefting11–16. Despite this success, the genetic variants identified so far collectively explain only a minor fraction of the total variance attributable to additive genetic effects, even though the heritability of CL/P among Europeans is more than 70%17–20. This has spurred renewed interest in investigating disease mechanisms other than fetal or maternal effects21. One example is parent-of-origin (PoO), where the effect of a particular allele in the offspring differs according to its parental origin22–24, and another is gene-environment interaction (GxE), where fetal effects differ across strata of environmental exposures25. Identifying GxE effects may not only provide new insights into the causes of CL/P, but may also provide an opportunity to intervene on environmental risk factors alone, particularly in subgroups of the population that are genetically more susceptible to these environmental effects.
Recently, we went one step further and developed new methods for a genome-wide screening for PoO interactions with environmental exposures (i.e., PoOxE) in the case-parent triad setting22. We applied the new methodology, implemented in the R-package Haplin26, to isolated cleft palate only (CPO)27, using genotypes and exposure data from the largest published GWAS dataset on case-parent triads of orofacial clefts11. Epidemiological and embryological findings have previously shown that CL/P and CPO may have distinct etiologies. Therefore, we used the same GWAS dataset and methodology to perform a genome-wide scan for PoOxE effects in the larger sample of isolated CL/P.
The study participants were mainly of Asian or European origin and were recruited as part of an international cleft collaboration11. Information was available on genotypes as well as maternal vitamin use, cigarette smoking and alcohol consumption in the periconceptional period (three months before and three months after pregnancy). The information on environmental exposures was based on interviews and questionnaires. More detailed characteristics of the study participants can be found in our recent work28.
Table 1 shows the distribution of the CL/P families according to ethnicity, triad completeness and maternal exposure. There were 1908 families in the pooled sample (5424 individuals in total), which included all the participants. Of these, 825 families were in the European sample, 1024 families were in the Asian sample, and 59 families were in the sample consisting of other ethnicities (Table 1). We performed three main sets of analyses on the following samples: All participants (denoted as “pooled analysis”), only Asians (“Asian analysis”), and only Europeans (“European analysis”). The 59 families with other ethnicities were not analyzed as a group due to the small sample size, but they were included in the pooled analysis. In the pooled and European analyses, we examined all exposures. As cigarette smoking and alcohol consumption were rare among Asian mothers, we were only able to conduct PoOxVitamin analyses for this ethnicity.
| Complete + incomplete triads | Total | Mother exposed (missing) | |||||
|---|---|---|---|---|---|---|---|
| Ethnicity | Individuals | Families | Individuals | Families | Alcohol | Smoking | Vitamin |
| European | 2024+310 | 670+155 | 2334 | 825 | 325 (8) | 249 (6) | 462 (98) |
| Asiana | 2670+268 | 890+134 | 2938 | 1024 | - | - | 142 (155) |
| Otherb | 102+ 50 | 34+ 25 | 152 | 59 | - | - | - |
| Pooled | 4796+628 | 1594+314 | 5424 | 1908 | 350 (22) | 284 (9) | 638 (255) |
aNo analyses of parent-of-origin interactions with alcohol (PoOxAlcohol) or parent-of-origin interactions with smoking (PoOxSmoke) were conducted for this group because of a lack of observations for these exposures. bOwing to the small sample size, no analysis of parent-of-origin interactions with environmental exposures (PoOxE) was conducted for this group. Note that a subset of the complete triads included more than one offspring. Incomplete triads are parent-offspring dyads.
Quality control for excluding single-nucleotide polymorphisms (SNPs) and individuals were conducted as described in Haaland et al. (2017)27. That is, we included SNPs with a missing call rate less than 5%, a minor allele frequency (MAF) greater than 5%, a p-value of less than 0.001 for the test for Hardy-Weinberg equilibrium presented by Wigginton et al. (2005)29, and a Mendelian error rate greater than 10%. Further, if two or more SNPs were in perfect linkage disequilibrium (r2=1) with each other, we only included one in the analyses. After applying these same criteria here, 341,191 were left for the current analyses from a total of 569,244 SNPs (Table 2).
| Total number of single-nucleotide polymorphisms (SNPs) | 569,244 |
| Criteria: | |
| Failed HWE test (p<0.001) | 173,955 |
| More than 5% missing calls | 1934 |
| MAF less than 5% | 61,167 |
| r2=1 with flanking SNPs | 2880 |
| Mendelian errors detected (>1%) | 349 |
| Number of SNPs remaining after quality controla | 341,191 |
For statistical analysis, we used the statistical software Haplin26, which is written in the R statistical programming language30. Haplin is based on log-linear modeling in a maximum likelihood framework and is well-suited for the analysis of offspring-parent triads. Because Haplin uses the expectation-maximization (EM) algorithm to account for missing parental genotypes26, we were able to include the 314 case-parent dyads in the analyses beside the complete triads (Table 1). Haplin also uses the EM algorithm to reconstruct haplotypes, which enabled haplotype analyses for different combinations of SNPs in the genes that showed a plausible PoOxE effect.
A detailed description of the method for PoOxE analysis has been provided in our previous works22,27,31. Briefly, PoOxE effects were calculated as follows:
1) Calculate the relative risk (RR) for an allele inherited from the mother (RRmat) and do the same for the father (RRpat).
2) Calculate the relative risk ratio (RRRPoO=RRmat/RRpat) between the RRs in (1). RRRPoO is thus an estimate of the parent-of-origin (PoO) effect.
3) Calculate RRRPoOxE as RRRPoO(Exposed)/RRRPoO(Unexposed), where RRRPoO(Exposed) and RRRPoO(Unexposed) are RRRPoO among triads with exposed and unexposed mothers.
Haplin uses a Wald test to test the null hypothesis of RRRPoOxE=1.
In order to control for multiple testing (one test for each of 341,191 SNPs), we obtained q-values using the false discovery rate (FDR) method described by Storey & Tibshirani (2003)32. Specifically, the q-values were calculated from the p-values with the R-function qvalue()33. A q-value of 0.2 corresponds to an FDR of 20%, which means that at least 80% of SNPs with a q-value less than 0.2 would be expected to be truly associated with the outcome. As in our previous work on isolated CPO27, we identified the top 20 SNPs for each of the analyses performed (see Results for details) and calculated relative risk ratios (RRRs) with 95% confidence intervals (95% CI). We paid more attention to a given gene if SNPs in that gene showed up multiple times in one set or across different sets of analyses. In accordance with recent recommendations by the American Statistical Association and others34,35, we did not consider a fixed p-value as a threshold for statistical significance.
To illustrate the general ability of the PoOxE analyses to detect true associations, power analyses for a wide range of PoOxE scenarios were performed using the Haplin function hapPowerAsymp(), as described in our recent works22,36.
We focused on the regions flanking SNPs in the most interesting genes and constructed regional plots based on R-scripts developed by the Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University and Novartis Institutes of BioMedical Research37. Such plots capture the extent of linkage disequilibrium between a lead SNP and neighboring SNPs, while also providing information on recombination patterns and the position of genes.
R-scripts used to conduct the statistical analyses and create figures are available (see Software availability)38.
To contextualize the findings, we searched for connections among a selection of genes in the STRING database39, as well as for enrichment of these genes in expression patterns using ExpressionAtlas40 and BGee (R package BgeeDB_2.10.0)41. Further, using Hetionet (Ver.1.0)42, we searched for indirect links between the genes highlighted by our analyses, the exposures and the phenotype (“cleft lip”). Hetionet is a heterogeneous network of various relationships among various data types, such as interactions between genes, or regulation of gene expression between a drug and a gene. The data used in Hetionet were carefully curated from 29 publicly available databases. To simplify the query output, the number of relationships between any two of the input query nodes (i.e., exposure, cleft lip, and the genes) was set to at most two. The exact queries together with their output are available (see Software availability)38.
The individual institutional review boards of the members of the International Cleft Consortium provided ethical approval, which can be found in the online supplementary material of the original publication43. Written informed consent was provided by all participating families. Please refer to the dbGaP database for more information.
For clarity, this section is structured as follows: We present the results of the PoOxE analyses of the pooled sample first (Table 3), followed by those of the European (Table 4) and Asian (Table 5) samples. We used the integrative database GeneCards and the gene-centric links therein to collate information on the genes in these tables. The 1000 Genomes browser was used to determine the chromosomal band location of a SNP. In the following sections, we focus on q-values, but all the corresponding p-values can also be found in Table 3–Table 5. Table 6 provides a reference for the full names of all the genes mentioned in Table 3–Table 5. Table 7 shows the results of the haplotype analyses of SNPs in the most interesting genes. Figure 1 and Figure 2 present visualizations of the results from the bioinformatics analyses, and regional plots for the most important regions from Table 3–Table 5 are shown in Figure 3 and Figure 4. Figure 5 illustrates power calculations to detect different PoOxE effects in single-SNP analyses under different parameters, such as different sample sizes and minor allele frequencies. Quantile-quantile (QQ) plots for each set of analyses are shown in Figure 6–Figure 8.
| Exposure | SNP | Chromosomal band locationa | P-value | Q-value | RRR (95% CI) | Gene symbolb | Sharedc |
|---|---|---|---|---|---|---|---|
| ALCOHOL | rs7964474 | 12p13.31 | 7.4e-06 | 0.99 | 0.34 (0.22-0.55) | ANO2 | |
| rs999783 | 16q23.3-q24.1 | 1.8e-05 | 0.99 | 2.63 (1.69-4.10) | MBTPS1 | ||
| rs4982619 | 14q11.2 | 2.1e-05 | 0.99 | 2.44 (1.62-3.68) | TRA | ||
| rs7945550 | 11p13 | 2.1e-05 | 0.99 | 2.46 (1.62-3.72) | EHF | Europe | |
| rs880813 | 2p12 | 2.5e-05 | 0.99 | 2.36 (1.58-3.51) | CTNNA2 | ||
| rs2280025 | 16q23.3-q24.1 | 2.7e-05 | 0.99 | 2.59 (1.66-4.03) | MBTPS1 | ||
| rs11584506 | 1q42.1 | 3.4e-05 | 0.99 | 0.39 (0.25-0.61) | NC | ||
| rs10897066 | 11q12.2 | 3.8e-05 | 0.99 | 2.29 (1.54-3.40) | ~MS4A5 and MS4A1 | ||
| rs2032442 | 14q11.2 | 3.9e-05 | 0.99 | 2.37 (1.57-3.59) | TRA | ||
| rs163684 | 12q14.1-q14.2 | 4.2e-05 | 0.99 | 3.23 (1.84-5.65) | PPM1H | ||
| rs8025763 | 15q26.3 | 5.6e-05 | 0.99 | 2.31 (1.54-3.47) | NC | ||
| rs13008096 | 2p15 | 6.1e-05 | 0.99 | 2.26 (1.52-3.36) | NC | ||
| rs4699228 | 4q24 | 6.2e-05 | 0.99 | 2.80 (1.69-4.63) | NC | ||
| rs2723057 | 4q24 | 6.2e-05 | 0.99 | 2.76 (1.68-4.54) | NC | ||
| rs7201659 | 16p12.3 | 6.5e-05 | 0.99 | 0.43 (0.29-0.65) | XYLT1 | ||
| rs2151225 | 9q21.3 | 6.8e-05 | 0.99 | 2.54 (1.61-4.02) | NC | ||
| rs7197476 | 16p12.3 | 6.9e-05 | 0.99 | 0.44 (0.29-0.66) | XYLT1 | ||
| rs2367283 | 9q21.3 | 7.0e-05 | 0.99 | 0.42 (0.28-0.65) | GPR98 | ||
| rs2914354 | 19q13.42 | 7.7e-05 | 0.99 | 0.47 (0.32-0.68) | ~VN1R4 | ||
| rs7209652 | 17p12 | 8.7e-05 | 0.99 | 0.45 (0.30-0.67) | LINC00670 | ||
| SMOKE | rs10097386 | 8q22.1 | 2.6e-06 | 0.57 | 2.86 (1.85-4.43) | NC | |
| rs2383162 | 9p21.3 | 8.5e-06 | 0.57 | 2.73 (1.75-4.24) | FOCAD | ||
| rs10738571 | 9p21.3 | 1.3e-05 | 0.57 | 2.67 (1.72-4.16) | FOCAD | ||
| rs7419201 | 1q43 | 1.4e-05 | 0.57 | 3.21 (1.90-5.44) | NC | Europe | |
| rs7541537 | 1q43 | 1.4e-05 | 0.57 | 2.52 (1.66-3.81) | NC | Europe | |
| rs7042192 | 9p21.3 | 1.5e-05 | 0.57 | 2.70 (1.72-4.22) | FOCAD | ||
| rs4977848 | 9p21.3 | 1.5e-05 | 0.57 | 2.71 (1.72-4.24) | FOCAD | ||
| rs7920088 | 10p14 | 1.6e-05 | 0.57 | 2.94 (1.80-4.81) | SFMBT2 | ||
| rs12740826 | 1q25.2 | 1.9e-05 | 0.57 | 0.35 (0.22-0.57) | NPHS2 | ||
| rs13173741 | 5q14.1 | 2.5e-05 | 0.57 | 2.51 (1.63-3.84) | NC | ||
| rs10757168 | 9p21.3 | 2.5e-05 | 0.57 | 2.60 (1.67-4.05) | FOCAD | ||
| rs8181543 | 11q22.3 | 2.6e-05 | 0.57 | 0.34 (0.21-0.56) | PDGFD | Europe | |
| rs168283 | 4q21.21 | 2.7e-05 | 0.57 | 0.36 (0.22-0.58) | FRAS1 | ||
| rs17408603 | 1p31.1 | 2.7e-05 | 0.57 | 3.19 (1.86-5.49) | NC | ||
| rs11624380 | 14q22.3 | 2.8e-05 | 0.57 | 0.37 (0.23-0.59) | PELI2 | ||
| rs2177971 | 8p21.2 | 2.8e-05 | 0.57 | 3.43 (1.93-6.11) | NC | Europe | |
| rs7943401 | 11q22.3 | 2.9e-05 | 0.57 | 0.34 (0.20-0.56) | PDGFD | ||
| rs3793861 | 10q21.2 | 3.0e-05 | 0.57 | 2.71 (1.70-4.34) | ANK3 | Europe | |
| rs4394682 | 1p36.13 | 3.4e-05 | 0.58 | 0.34 (0.20-0.57) | ~CAPZB | ||
| rs7087489 | 10q21.2 | 3.5e-05 | 0.58 | 2.69 (1.68-4.30) | ANK3 | Europe | |
| VITAMIN | rs2302304 | 19p13.3 | 1.3e-06 | 0.46 | 3.12 (1.97-4.94) | TJP3 | |
| rs2689128 | 1q43 | 4.2e-06 | 0.71 | 3.28 (1.98-5.43) | NC | Europe | |
| rs9572250 | 13q21.33 | 7.8e-06 | 0.88 | 0.44 (0.31-0.63) | KLHL1 | ||
| rs4875398 | 8p23.2 | 1.4e-05 | 0.99 | 2.08 (1.49-2.89) | CSMD1 | ||
| rs3909551 | 13q21.33 | 1.7e-05 | 0.99 | 0.46 (0.32-0.65) | KLHL1 | ||
| rs9371494 | 6q25.1 | 2.4e-05 | 0.99 | 2.23 (1.54-3.24) | MTHFD1L | ||
| rs8101981 | 19p13.12 | 2.9e-05 | 0.99 | 0.48 (0.34-0.68) | LINC00905 | Europe | |
| rs7939975 | 11.p12 | 3.6e-05 | 0.99 | 2.08 (1.47-2.94) | NC | ||
| rs10495767 | 2p23.2 | 3.6e-05 | 0.99 | 2.28 (1.54-3.36) | NC | ||
| rs11673884 | 2q36.3 | 4.2e-05 | 0.99 | 0.51 (0.37-0.70) | ~SLC19A3 | ||
| rs6489630 | 12p13.31 | 4.6e-05 | 0.99 | 2.23 (1.52-3.28) | NTF3 | ||
| rs3815311 | 17p12 | 5.3e-05 | 0.99 | 3.19 (1.82-5.59) | ARHGAP44 | Europe | |
| rs358017 | 3p21.1-p14.3 | 5.4e-05 | 0.99 | 2.25 (1.52-3.34) | ~CACNA2D3 | ||
| rs7082286 | 10q21.1 | 5.8e-05 | 0.99 | 4.03 (2.04-7.96) | NC | ||
| rs921743 | 10p13 | 6.0e-05 | 0.99 | 2.18 (1.49-3.19) | RSU1 | ||
| rs10764037 | 10p12.31 | 6.3e-05 | 0.99 | 0.50 (0.36-0.70) | MALRD1 | ||
| rs8112256 | 19p13.11 | 6.8e-05 | 0.99 | 2.13 (1.47-3.10) | FAM129C | ||
| rs4569521 | 2q21.1 | 8.1e-05 | 0.99 | 0.42 (0.27-0.65) | ARHGEF4 | ||
| rs6830509 | 4q28 | 8.7e-05 | 0.99 | 1.96 (1.40-2.73) | NC | ||
| rs9503155 | 6p25.3 | 8.8e-05 | 0.99 | 0.49 (0.34-0.70) | GMDS-AS1 |
aThe 1000 Genomes browser was used to determine the chromosomal band location of a SNP. bIf a SNP is located within a gene itself, the gene symbol is provided (the full names of the genes are provided in Table 6). SNPs located within 40 kb of a gene have the prefix ‘~’, and those not located within a 40 kb-distance of a gene are denoted as NC (for ‘not close’). Note that pseudogenes and non-coding RNAs are excluded. cShared: Also featured in Table 4 or Table 5. SNP, single-nucleotide polymorphism; RRR, relative risk ratio; CI, confidence interval; NC, not close.
| Exposure | SNP | Chromosomal band locationa | P-value | Q-value | RRR (95% CI) | Gene symbolb | Sharedc |
|---|---|---|---|---|---|---|---|
| ALCOHOL | rs10496410 | 2q12 | 7.5e-07 | 0.15 | 6.04 (2.96-12.32) | NC | |
| rs7579926 | 2q12 | 9.3e-07 | 0.15 | 5.95 (2.92-12.13) | NC | ||
| rs2294035 | 8p23.3 | 2.9e-06 | 0.32 | 0.31 (0.19-0.51) | ARHGEF10 | ||
| rs6975650 | 7q33 | 1.1e-05 | 0.76 | 0.31 (0.19-0.52) | NC | ||
| rs4876274 | 8p23.3 | 1.3e-05 | 0.76 | 2.99 (1.83-4.90) | ARHGEF10 | ||
| rs2245225 | 12q14 | 1.4e-05 | 0.76 | 3.46 (1.98-6.05) | NC | ||
| rs927318 | 9p24.2 | 2.0e-05 | 0.76 | 0.36 (0.22-0.57) | GLIS3 | ||
| rs10735337 | 12q23.1 | 2.0e-05 | 0.76 | 0.36 (0.23-0.58) | CCDC38 | ||
| rs6427247 | 1q24 | 2.1e-05 | 0.76 | 2.87 (1.77-4.67) | NC | ||
| rs12669493 | 7p21.1 | 2.4e-05 | 0.79 | 3.11 (1.84-5.26) | LRRC72 | ||
| rs13255561 | 8p23.3 | 3.6e-05 | 0.88 | 0.30 (0.17-0.53) | DLGAP2 | ||
| rs12242535 | 10q21.2 | 3.9e-05 | 0.88 | 3.94 (2.05-7.57) | NC | ||
| rs943881 | 14q32.2 | 4.3e-05 | 0.88 | 0.36 (0.22-0.59) | CYP46A1 | ||
| rs10491327 | 5q34 | 4.4e-05 | 0.88 | 0.28 (0.15-0.52) | NC | ||
| rs7945550 | 11p13 | 4.5e-05 | 0.88 | 2.82 (1.71-4.64) | EHF | Pooled | |
| rs7232492 | 18p11.31 | 5.3e-05 | 0.88 | 0.27 (0.15-0.51) | DLGAP1 | ||
| rs11242213 | 5q31.1 | 5.4e-05 | 0.88 | 4.71 (2.22-10.00) | UBE2B | ||
| rs34352212 | 5q34 | 6.0e-05 | 0.88 | 0.32 (0.18-0.55) | NC | ||
| rs1990185 | 17q24 | 6.1e-05 | 0.88 | 3.16 (1.80-5.54) | NC | ||
| rs521419 | 17p12 | 6.5e-05 | 0.88 | 2.80 (1.69-4.64) | NC | ||
| SMOKE | rs10763707 | 10p12.1-p11.23 | 1.5e-06 | 0.20 | 4.08 (2.30-7.23) | LYZL1 | |
| rs7541537 | 1q43 | 2.0e-06 | 0.20 | 3.31 (2.02-5.43) | NC | Pooled | |
| rs7419201 | 1q43 | 2.1e-06 | 0.20 | 4.77 (2.50-9.11) | NC | Pooled | |
| rs3793861 | 10q21.2 | 2.6e-06 | 0.20 | 3.67 (2.13-6.32) | ANK3 | Pooled | |
| rs7087489 | 10q21.2 | 3.1e-06 | 0.20 | 3.63 (2.11-6.25) | ANK3 | Pooled | |
| rs814518 | 19q13.2 | 4.5e-06 | 0.25 | 3.35 (2.00-5.62) | SHKBP1 | ||
| rs4693142 | 4q21.3 | 6.4e-06 | 0.30 | 0.26 (0.15-0.47) | MAPK10 | ||
| rs4454616 | 10p14 | 9.2e-06 | 0.38 | 3.06 (1.86-5.00) | NC | ||
| rs2904096 | 4q21.3 | 1.2e-05 | 0.40 | 0.27 (0.15-0.49) | MAPK10 | ||
| rs2290682 | 19q13.2 | 1.3e-05 | 0.40 | 3.22 (1.90-5.44) | SHKBP1 | ||
| rs6532013 | 4q22 | 1.4e-05 | 0.40 | 3.04 (1.84-5.02) | NC | ||
| rs1868368 | 8q24.2 | 2.2e-05 | 0.61 | 0.29 (0.17-0.52) | NC | ||
| rs2177971 | 8p21.2 | 2.6e-05 | 0.64 | 4.02 (2.10-7.68) | NC | Pooled | |
| rs6807522 | 3q22.1 | 3.4e-05 | 0.67 | 2.91 (1.75-4.81) | TMEM108 | ||
| rs17604550 | 15q25.3 | 3.6e-05 | 0.67 | 0.33 (0.20-0.56) | AGBL1 | ||
| rs12883776 | 14q22.3 | 3.7e-05 | 0.67 | 0.34 (0.20-0.57) | PELI2 | ||
| rs7234787 | 18q21.1 | 3.8e-05 | 0.67 | 0.22 (0.11-0.45) | ZBTB7C | ||
| rs8181543 | 11q22.3 | 3.8e-05 | 0.67 | 0.28 (0.16-0.52) | PDGFD | Pooled | |
| rs4310561 | 10q21.2 | 4.0e-05 | 0.67 | 2.90 (1.75-4.83) | ANK3 | ||
| rs3800036 | 6p25.3 | 4.1e-05 | 0.67 | 0.35 (0.22-0.58) | GMDS | ||
| VITAMIN | rs2689128 | 1q43 | 2.2e-06 | 0.44 | 4.82 (2.52-9.25) | NC | Pooled |
| rs2237360 | 7p15.1 | 4.0e-06 | 0.44 | 0.29 (0.18-0.50) | CREB5 | ||
| rs7793050 | 7p21 | 4.0e-06 | 0.44 | 3.94 (2.20-7.05) | RPA3-AS1 | ||
| rs7766106 | 6q22.33 | 6.4e-06 | 0.53 | 0.31 (0.19-0.52) | RSPO3 | ||
| rs2809964 | 1p36.11 | 1.2e-05 | 0.65 | 3.05 (1.85-5.03) | ~RCAN3, NCMAP and RPL26P8 | ||
| rs3859121 | 16q12.1 | 1.2e-05 | 0.65 | 0.16 (0.07-0.37) | N4BP1 | ||
| rs1092733 | 3p26 | 2.4e-05 | 0.87 | 0.32 (0.19-0.54) | NC | ||
| rs7559678 | 2q11.2 | 2.7e-05 | 0.87 | 0.35 (0.22-0.57) | VWA3B | ||
| rs2366837 | 5p13.2 | 3.0e-05 | 0.87 | 0.35 (0.21-0.57) | NC | ||
| rs10084852 | 4q28.3 | 3.0e-05 | 0.87 | 7.26 (2.86-18.45) | PCDH10 | ||
| rs6446389 | 4p16.2 | 4.2e-05 | 0.87 | 3.38 (1.89-6.06) | EVC2 | ||
| rs2242909 | 21q22.1 | 4.2e-05 | 0.87 | 2.92 (1.75-4.87) | NC | ||
| rs595536 | 1q42.2 | 4.4e-05 | 0.87 | 3.21 (1.83-5.60) | ~SIPA1L2 | ||
| rs6726527 | 2q37.1 | 4.5e-05 | 0.87 | 0.24 (0.12-0.48) | ~SP140 and SP140L | ||
| rs12733019 | 1p32.1 | 4.8e-05 | 0.87 | 0.24 (0.12-0.47) | NC | ||
| rs8101981 | 19p13.12 | 4.8e-05 | 0.87 | 0.34 (0.21-0.58) | LINC00905 | Pooled | |
| rs17793145 | 8p22 | 4.9e-05 | 0.87 | 3.39 (1.88-6.12) | DLC1 | ||
| rs4973310 | 2q37.1 | 5.1e-05 | 0.87 | 0.24 (0.12-0.48) | ~SP140 and SP140L | ||
| rs3815311 | 17p12 | 5.5e-05 | 0.87 | 4.57 (2.18-9.57) | ARHGAP44 | Pooled | |
| rs8072885 | 17q25.3 | 5.5e-05 | 0.87 | 0.22 (0.10-0.46) | RBFOX3 |
aThe 1000 Genomes browser was used to determine the chromosomal band location of a SNP. bIf a SNP is located within a gene itself, the gene symbol is provided (the full names of the genes are provided in Table 6). SNPs located within 40 kb of a gene have the prefix ‘~’, and those not located within a 40 kb-distance of a gene are denoted as NC (for ‘not close’). Note that pseudogenes and non-coding RNAs are excluded. cShared: Also featured in Table 3 or Table 5. SNP, single-nucleotide polymorphism; RRR, relative risk ratio; CI, confidence interval; NC, not close.
| SNP | Chromosomal band locationa | P-value | Q-value | RRR (95% CI) | Gene symbola |
|---|---|---|---|---|---|
| rs1889976 | 1q25.3 | 8.8e-06 | 0.86 | 3.88 (2.13-7.05) | SWT1 |
| rs259395 | 6q24.3 | 1.1e-05 | 0.86 | 0.23 (0.12-0.45) | ADGB |
| rs10798004 | 1q25.3 | 1.5e-05 | 0.86 | 3.70 (2.04-6.68) | ~IVNS1ABP and SWT1 |
| rs12431484 | 14q11.2 | 2.2e-05 | 0.86 | 0.24 (0.12-0.46) | TRA |
| rs10518981 | 15q15.3-q21.1 | 2.3e-05 | 0.86 | 0.22 (0.11-0.45) | ~CTDSPL2 and EIF3J-AS1 and EIF3J |
| rs1940698 | 11q23.2 | 2.4e-05 | 0.86 | 0.21 (0.10-0.43) | NCAM1 |
| rs171477 | 21q21 | 2.5e-05 | 0.86 | 0.23 (0.12-0.46) | C21orf91-OT1 |
| rs9862866 | 3p14.1 | 3.1e-05 | 0.86 | 0.24 (0.12-0.47) | ~RPL21P41 |
| rs865585 | 6p21.1 | 3.6e-05 | 0.86 | 0.19 (0.09-0.42) | NC |
| rs17591732 | 11q23.2 | 3.8e-05 | 0.86 | 0.22 (0.11-0.45) | NCAM1 |
| rs12630106 | 3q13.1 | 5.6e-05 | 0.86 | 3.56 (1.92-6.61) | NC |
| rs7316350 | 12q15 | 6.0e-05 | 0.86 | 0.22 (0.10-0.46) | NC |
| rs7336296 | 13q31 | 6.1e-05 | 0.86 | 3.39 (1.87-6.17) | NC |
| rs1499916 | 2q22 | 6.4e-05 | 0.86 | 0.22 (0.10-0.46) | NC |
| rs7153574 | 14q11.2 | 6.5e-05 | 0.86 | 0.26 (0.13-0.50) | TRA |
| rs6439772 | 3q22 | 7.0e-05 | 0.86 | 0.26 (0.14-0.51) | NC |
| rs1348564 | 3q22 | 7.1e-05 | 0.86 | 0.27 (0.14-0.52) | NC |
| rs2360838 | 11p15.4 | 7.3e-05 | 0.86 | 3.37 (1.85-6.14) | ~OR10A3 and NLRP10 and OR10A6 |
| rs12204808 | 6q14.1 | 7.3e-05 | 0.86 | 4.63 (2.17-9.87) | IMPG1 |
| rs1407555 | 1q25.3 | 7.5e-05 | 0.86 | 3.30 (1.83-5.96) | TRMT1L |
aThe 1000 Genomes browser was used to determine the chromosomal band location of a SNP. bIf a SNP is located within a gene itself, the gene symbol is provided (the full names of the genes are provided in Table 6). SNPs located within 40 kb of a gene have the prefix ‘~’, and those not located within a 40 kb-distance of a gene are denoted as NC (for ‘not close’). Note that pseudogenes and non-coding RNAs (ncRNA) are excluded. There is no column for “shared” here, as none of these SNPs featured among those listed in Table 3 or Table 4. SNP, single-nucleotide polymorphism; RRR, relative risk ratio; CI, confidence interval; NC, not close.
| Gene name | SNP/haplotype | aTarget allele/Reference | bFrequency | Effect type | cRRR (95% CI) | p-value |
|---|---|---|---|---|---|---|
| ANK3 | rs3793861 | c/G | 0.30 | Child | 1.06 (0.91-1.24) | 0.45 |
| GxSmoke | 1.20 (0.83-1.60) | 0.41 | ||||
| PoO | 1.39 (1.09-1.76) | 0.007 | ||||
| PoOxSmoke | 3.67 (2.13-6.32) | 2.6e-6 | ||||
| rs7087489 | t/A | 0.30 | Child | 1.06 (0.91-1.24) | 0.44 | |
| GxSmoke | 1.10 (0.82-1.60) | 0.42 | ||||
| PoO | 1.40 (1.10-1.77) | 0.006 | ||||
| PoOxSmoke | 3.63 (2.11-6.25) | 3.1e-6 | ||||
| rs4310561 | a/T | 0.34 | Child | 1.12 (0.97-1.24) | 0.13 | |
| GxSmoke | 1.20 (0.87-1.60) | 0.27 | ||||
| PoO | 1.31 (1.03-1.64) | 0.02 | ||||
| PoOxSmoke | 2.90 (1.75-4.83) | 4.0e-5 | ||||
| rs3793861- rs7087489 | c-t/G-A | 0.30 | Child | 1.07 (0.92-1.24) | 0.39 | |
| GxSmoke | 1.10 (0.81-1.60) | 0.45 | ||||
| PoO | 1.38 (1.08-1.75) | 0.008 | ||||
| PoOxSmoke | 3.71 (2.16-6.39) | 2.2e-6 | ||||
| rs7087489- rs4310561 | A-a/A-T | 0.04 | Child | 1.32 (0.95-1.83) | 0.10 | |
| GxSmoke | 1.50 (0.71-3.10) | 0.29 | ||||
| PoO | 0.93 (0.60-1.45) | 0.74 | ||||
| PoOxSmoke | 1.57 (0.62-3.96) | 0.34 | ||||
| t-a/A-T | 0.30 | Child | 1.09 (0.94-1.28) | 0.26 | ||
| GxSmoke | 1.20 (0.83-1.60) | 0.37 | ||||
| PoO | 1.34 (1.06-1.70) | 0.02 | ||||
| PoOxSmoke | 3.65 (2.13-6.28) | 2.7e-6 | ||||
| rs3793861- rs7087489- rs4310561 | G-A-a/G-A-T | 0.04 | Child | 1.32 (0.95-1.83) | 0.10 | |
| GxSmoke | 1.50 (0.71-3.10) | 0.29 | ||||
| PoO | 0.93 (0.60-1.45) | 0.75 | ||||
| PoOxSmoke | 1.56 (0.62-3.94) | 0.35 | ||||
| c-t-a/G-A-T | 0.30 | Child | 1.09 (0.94-1.28) | 0.26 | ||
| GxSmoke | 1.20 (0.83-1.60) | 0.37 | ||||
| PoO | 1.35 (1.06-1.71) | 0.01 | ||||
| PoOxSmoke | 3.62 (2.10-6.21) | 3.3e-6 | ||||
| ARHGEF10 | rs2294035 | a/T | 0.49 | Child | 0.94 (0.82-1.08) | 0.38 |
| GxAlcohol | 1.20 (0.87-1.50) | 0.32 | ||||
| PoO | 0.95 (0.75-1.20) | 0.67 | ||||
| PoOxAlcohol | 0.32 (0.19-0.51) | 2.9e-6 | ||||
| rs4876274 | t/A | 0. 47 | Child | 1.04 (0.90-1.20) | 0.57 | |
| GxAlcohol | 0.90 (0.67-1.20) | 0.47 | ||||
| PoO | 1.02 (0.80-1.29) | 0.90 | ||||
| PoOxAlcohol | 2.99 (1.83-4.90) | 1.3e-5 | ||||
| rs2294035-rs4876274 | T-A/a-A | 0.04 | Child | 1.15 (0.80-1.68) | 0.44 | |
| GxAlcohol | 0.73 (0.33-1.60) | 0.45 | ||||
| PoO | 1.41 (0.85-2.37) | 0.19 | ||||
| PoOxAlcohol | 1.56 (0.49-4.93) | 0.45 | ||||
| T-t/a-A | 0.47 | Child | 1.04 (0.90-1.20) | 0.63 | ||
| GxAlcohol | 0.90 (0.67-1.20) | 0.46 | ||||
| PoO | 1.00 (0.80-1.27) | 0.98 | ||||
| PoOxAlcohol | 3.20 (1.97-5.21) | 2.8e-6 |
aEffect allele or haplotype against the reference. Lowercase indicates the minor allele at the SNP. bMinor allele frequency for a given SNP. In haplotype analyses, this corresponds to the frequencies of haplotypes other than the reference. cRR for child effects; RRR for GxSmoke or GxAlcohol, PoO and PoOxSmoke or PoOxAlcohol. All p-values <0.05 are highlighted in bold. Note that in a two-SNP-haplotype, there are four possible combinations, and in a three-SNP-haplotype there are eight. However, only two or three of these combinations were actually observed in the data. SNP, single nucleotide polymorphism; RRR, relative risk ratio; RR, risk ratio; CI, confidence interval; PoO, parent-of-origin; GxSmoke, gene-smoking interaction; GxAlcohol, gene-alcohol interaction; PoOxAlcohol, parent-of-origin interactions with alcohol; PoOxSmoke, parent-of-origin interactions with smoking.
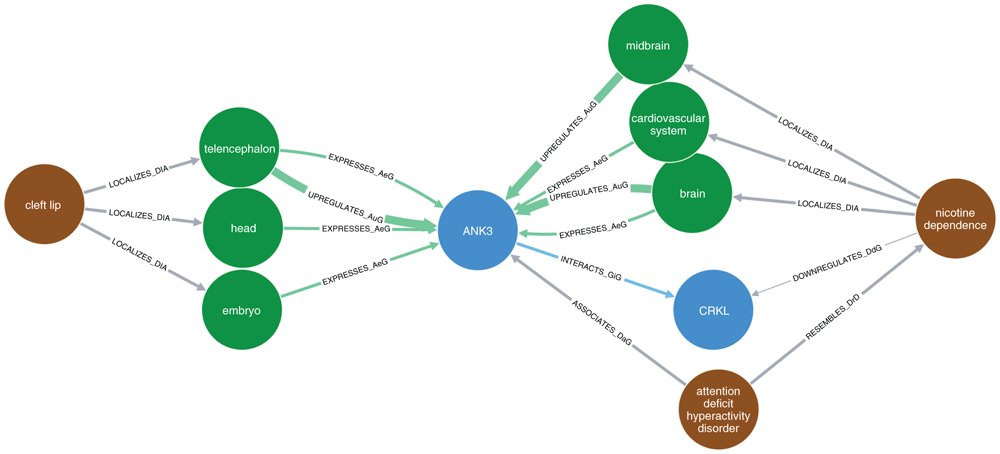
The brown nodes represent diseases, blue nodes show genes/proteins, and green nodes represent organs (anatomy). Each arrow represents a specific relationship between nodes: “LOCALIZES_DiA” = disease was found to be localized in an anatomy (organ); “EXPRESSES_AuG”, “UPREGULATES_AuG”, “DOWNREGULATES_AuG” mean that the gene is expressed, upregulated, or downregulated in the anatomy (organ), respectively; “INTERACTS_GiG” means that the two genes were found to interact with each other (physically, as proteins); “ASSOCIATES_DaG” means that the gene was found to be associated with the disease; “RESEMBLES_DrD” means that the two diseases were found to occur significantly more often together in MEDLINE articles than would be expected by chance alone. Note that in this setting, the term "disease" includes any adverse medical condition, like syndromes, mental disorders, congenital anomalies, and so on.
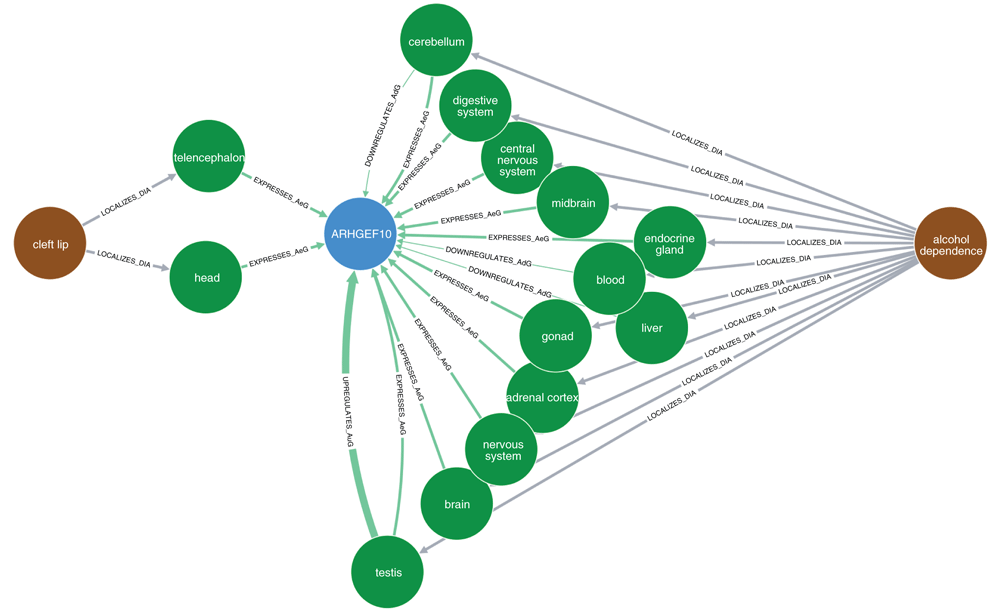
The brown nodes represent diseases, blue nodes show genes/proteins, and green nodes represent organs (anatomy). Each arrow represents a specific relationship between nodes: “LOCALIZES_DiA” = disease was found to be localized in an anatomy (organ); “EXPRESSES_AuG”, “UPREGULATES_AuG”, “DOWNREGULATES_AuG” mean that the gene is expressed, upregulated, or downregulated in the anatomy (organ), respectively; “INTERACTS_GiG” means that the two genes were found to interact with each other (physically, as proteins); “ASSOCIATES_DaG” means that the gene was found to be associated with the disease; “RESEMBLES_DrD” means that the two diseases were found to occur significantly more often together in MEDLINE articles than would be expected by chance alone. Note that in this setting, the term "disease" includes any adverse medical condition, like syndromes, mental disorders, congenital anomalies, and so on.

The plot provides information on the recombination rate and linkage disequilibrium between the lead SNP (blue diamond) and other SNPs in the region.
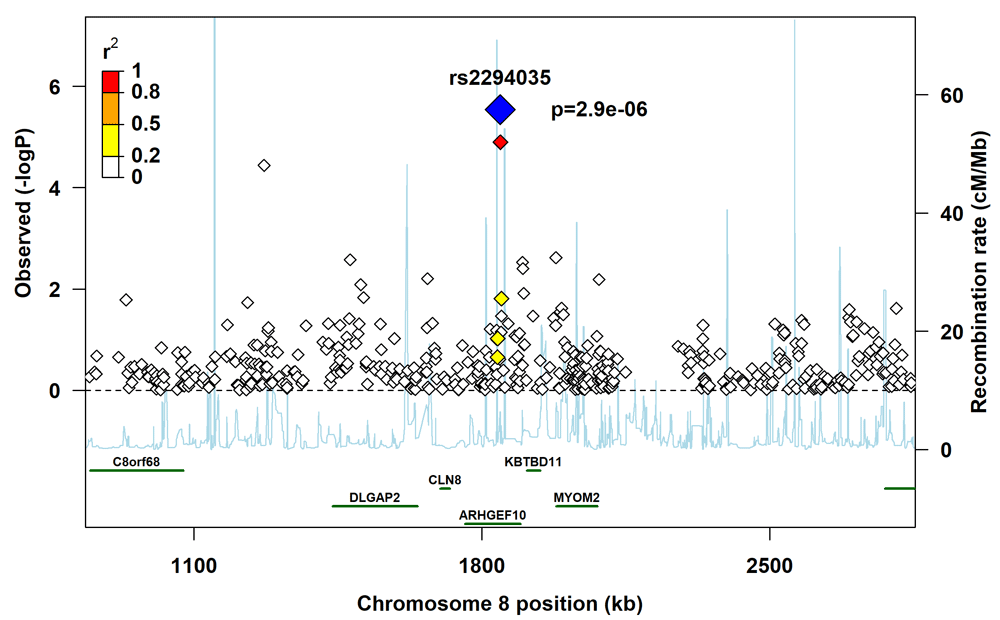
The plot provides information on the recombination rate and linkage disequilibrium between the lead SNP (blue diamond) and other SNPs in the region.
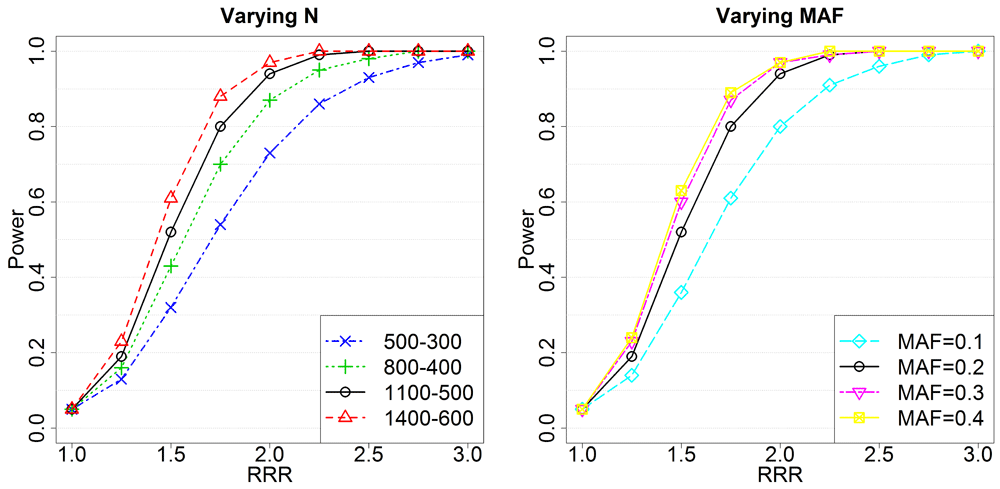
Left panel: Setting the minor allele frequency to 0.2 while varying the number of unexposed and exposed triads (unexposed-exposed). Right panel: Setting the number of unexposed and exposed triads to 1100 and 500, respectively, while varying the minor allele frequency. In all analyses, the significance level was 0.05. We varied the maternal RR in exposed triads, so that RRR=RRmat(Exposed). The black curve is the same in both panels because of shared parameters. RRR, relative risk ratio; RRmat, relative risk for an allele inherited from the mother; MAF, minor allele frequency.
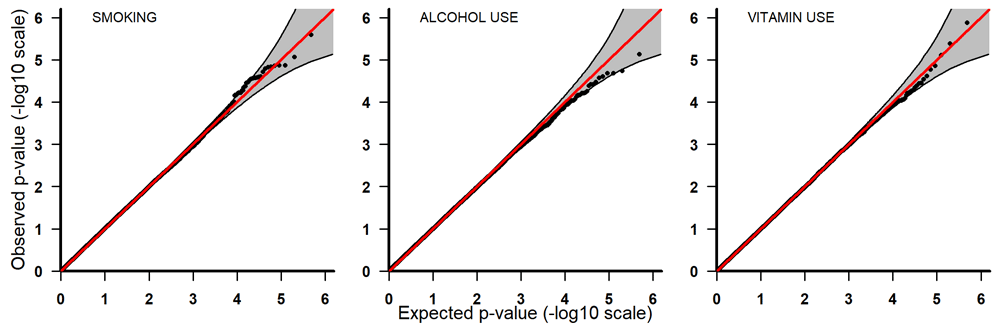
Q-Q plots for PoOxSmoke (left) PoOxAlcohol (middle) and PoOxVitamin (right) with 95% pointwise confidence bands. Q-Q, quantile-quantile; PoOxAlcohol, parent-of-origin interactions with alcohol; PoOxSmoke, parent-of-origin interactions with smoking; PoOxVitamin, parent-of-origin interactions with vitamins.
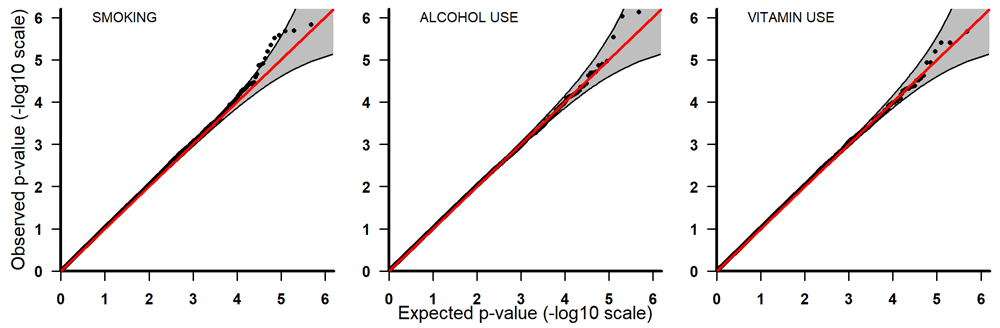
Q-Q plots for PoOxSmoke (left) PoOxAlcohol (middle) and PoOxVitamin (right) with 95% pointwise confidence bands. Q-Q, quantile-quantile; PoOxAlcohol, parent-of-origin interactions with alcohol; PoOxSmoke, parent-of-origin interactions with smoking; PoOxVitamin, parent-of-origin interactions with vitamins.
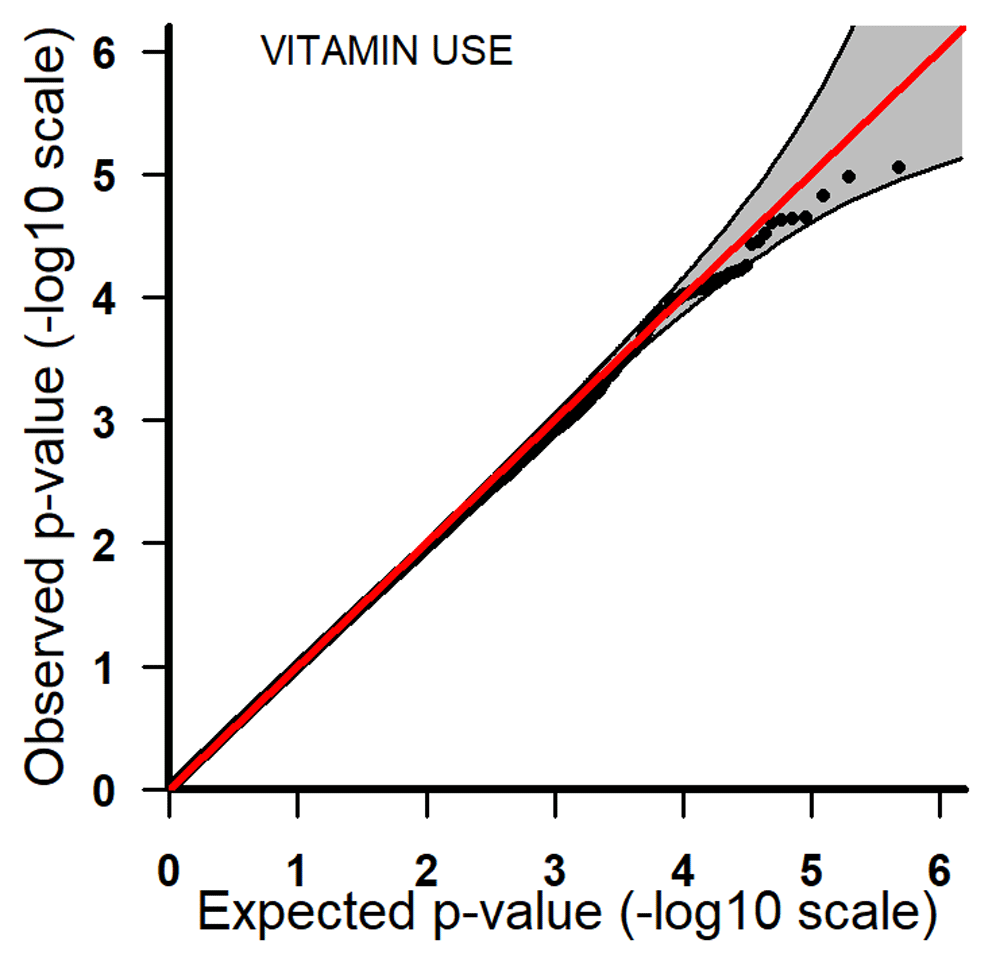
Q-Q plots for PoOxVitamin with 95% pointwise confidence bands. Q-Q, quantile-quantile; PoOxVitamin, parent-of-origin interactions with vitamins.
All the top 20 SNPs in the PoOxAlcohol analysis had the same q-value of 0.99 and are therefore not considered here as they are probably false positives (Table 3). All the SNPs in the PoOxSmoke analysis had q-values of around 0.6. Even though these q-values are still quite large, they indicate that around 40% of the SNPs are potentially true PoOxE associations. Among the top 20 SNPs in the PoOxSmoke analysis, two are in the gene for ‘Focadhesin’ (FOCAD), two are in ‘Platelet derived growth factor D’ (PDGFD), two are in ‘Ankyrin 3’ (ANK3), and one is in ‘Fraser syndrome 1’ (FRAS1). Note that associations with PDGFD and ANK3 were also detected in the European analyses (see below). In the PoOxVitamin analysis, only three SNPs had q-values below 0.99, and none of the genes linked to these SNPs have previously been associated with orofacial clefts.
Among the SNPs with the lowest q-values in the PoOxAlcohol analysis, rs2294035 (q=0.32, p=2.9e-6) and rs4876274 (q=0.76, p=1.3e-5) are in ‘Rho guanine nucleotide exchange factor 10’ (ARHGEF10; GeneCards identifier [GCID]: GC08P001823) (Table 4). The remaining SNPs had q-values above 0.76 and are not considered any further. ARHGEF10 has not previously been linked with orofacial clefts. In the PoOxSmoke analysis, three of the SNPs were in ANK3 (rs3793861: q=0.20, p=2.6e-6; rs7087489: q=0.20, p=3.1e-6; and rs4310561: q=0.67, p=4.0e-5). PoOxE effects in ANK3 were also detected in the analysis of the pooled sample above. To our knowledge, ANK3 has not previously been linked with orofacial clefts, and the same applies to SNP rs10763707 in ‘Lysosome like 1’ (LYZL1; GCID: GC10P029297), which had a q-value of 0.20. In the PoOxVitamin analysis, several of the SNPs shared the same q-value of 0.87 and are not considered any further. The top six SNPs had q-values of 0.44-0.65. Again, none of these genes appear to have any previous connections to clefting. For example, ‘cAMP responsive element binding protein 5’ (CREB5; GCID: GC07P028305) and its network of genes are involved in colorectal cancer44, while ‘R-Spondin 3’ (RSPO3; GCID: GC06P127118) is implicated in tumor development. That said, Park and co-workers reported that RSPO3 acts as an agonist in the canonical Wnt/β-catenin signaling45, a pathway known to be implicated in a wide range of developmental processes, including craniofacial development and homeostasis46–49.
In the only analysis possible for this ethnic group (PoOxVitamin), all the SNPs had the same q-value of 0.86 (Table 5). They are thus most likely to be false positives and will not be considered any further.
We chose to focus here on the PoOxE effects detected with SNPs in ANK3 and ARHGEF10. As mentioned above, ANK3 showed up several times among the top PoOxSmoke hits both in the pooled and European analyses, and strong signals for SNPs in ARHGEF10 were detected twice in the European analysis of PoOxAlcohol. We conducted stratified analyses of the effect of the child’s allele, GxE effects, PoO effects, and PoOxE effects for each SNP and haplotype (with haplotypes analyzed both in two-SNP and three-SNP combinations) in these two genes (Table 7). Specifically, we analyzed rs3793861, rs7087489 and rs4310561 in ANK3 that showed PoOxSmoke effects in the European sample, and rs2294035 and rs4876274 in ARHGEF10 that showed PoOxAlcohol effects in the same sample (Table 4). The results did not show any child effects or GxSmoke effects for single SNPs in ANK3. By contrast, the p-values were low for all three SNPs in the PoO analyses or PoOxSmoke analyses. This was also the case with the ‘t-a’ allele in the two-SNP combination rs3793861-rs7087489 and the ‘c-t-a’ allele in the three-SNP-combination rs3793861-rs7087489-rs4310561. The other alleles were only associated with PoOxSmoke effects.
For ARHGEF10, we analyzed the two SNPs that showed PoOxAlcohol effects in the European sample (rs2294035 and rs4876274) but did not discover any effects in either single-SNP or haplotype analyses.
Because of their low q-values, the genes appearing in the PoOxSmoke and PoOxAlcohol analyses in Table 4 were selected for further analyses using the STRING database, ExpressionAtlas and BGee. However, none of the searches for direct links among the genes yielded any evidence to explain why those genes appeared in our results together.
Regarding the indirect relationships, these are visualized in Figure 1 for relationships between ANK3 and cleft lip (Disease Ontology ID [DOID]: 9296), and, simultaneously, between ANK3 and nicotine dependence (DOID: 0050742). As Hetionet does not include information about smoking, we chose “nicotine dependence” as a proxy. ANK3 is connected to nicotine dependence through several nodes, two of which are particularly noteworthy. First, ANK3 has been reported to be strongly associated with attention-deficit/hyperactivity disorder (ADHD)50, and a connection between ADHD and nicotine dependence has been reported. The connectiom was calculated based on articles listed in MEDLINE, where this pair of conditions co-occured significantly more frequently than would be expected by chance51,52. The second path goes through the gene ‘CRK Like Proto-Oncogene, Adaptor Protein’ (CRKL). It interacts with ANK3 and is downregulated in nicotine dependence. Furthermore, ANK3 is expressed in the telencephalon (the most highly developed part of the forebrain), the embryo, and the head all of which are all relevant to CL/P.
Figure 2 shows the indirect relationships between cleft lip, ARHGEF10 and alcohol dependence. Like nicotine dependence in the above analyses, alcohol dependence (DOID: 0050741) was used here as a proxy for maternal alcohol consumption. The only relationships found between ARHGEF10 and cleft lip are the expression of ARHGEF10 in the head and telencephalon. By contrast, there were twelve different relationships between ARHGEF10 and alcohol dependence. However, there were no shared paths connecting cleft lip and alcohol dependence via any of the 12 organs.
The regional plot for rs3793861 (Figure 3) shows that several SNPs in ANK3 that were not in linkage disequilibrium with rs3793861 had p-values in the range 10-4 to 10-3, which lends support to either ANK3 itself or genes in its vicinity influencing the risk of clefting. However, we did not observe a similar pattern in the regional plot for rs2294035 (Figure 4).
Figure 5 shows that the power does not increase appreciably when the minor allele frequency increases beyond 0.2. However, there is a lot to be gained by increasing the sample size from 500 unexposed and 300 exposed (European, smoke/alcohol) to 1400-600 (pooled, vitamin). Further, the RRRs in the plots are based on changing only the effect of the maternal allele in the exposed triads. The same RRRs could have been achieved in a number of ways, which complicates the interpretation of the RRRs in Table 3–Table 5. Still, if a strong effect is detected with a SNP in a gene, this strengthens the case for its contribution to clefting.
The main aim of this paper was to identify genome-wide PoOxE effects in the larger sample of isolated CL/P, based on the same methodology and GWAS dataset we had previously used in a similar analysis of the smaller sample of isolated CPO27. As with the CPO study, the current analyses benefitted from being based on the largest available GWAS dataset of case-parent triads of orofacial clefts to date. Moreover, data were available for two major ethnicities, European and Asian, which is useful in assessing the generalizability of the findings across different ethnic groups. In the current dataset, however, very few of the Asian mothers reported smoking cigarettes or consuming alcohol during the periconceptional period, thus preventing a comparison of PoOxE effects for these exposures across these two ethnic groups. This is a common impediment to GxE studies, where the number of exposed individuals needs to be large enough for a meaningful analysis53.
A possible mechanism for a PoO effect is genomic imprinting21. This occurs when DNA methylation in the germline causes the expression of alleles to be silenced depending on their parental origin. Maternal environmental exposures that affect methylation patterns may also affect paternally and maternally inherited alleles differently. Furthermore, a PoO effect that is not affected by an environmental exposure will not be detected in our PoOxE analysis. Hence, a detected PoOxE effect may have a better chance than a PoO or GxE effect of uncovering a true causal relationship involving genomic imprinting.
Relying on the q-values for assessing the false positive rate, our analyses detected possible PoOxSmoke effects with SNPs in LYZL1, ANK3, PDGFD, FOCAD and FRAS1, and possible PoOxAlcohol effects with two SNPs in ARHGEF10. Without formal validation in a comparable and independent replication cohort, it would be premature to accept these associations as true PoOxE effects. Not having previously been linked with orofacial clefts does not necessarily imply that the identified gene is not relevant for clefting. This applies to several genes in our analyses; for example, SHKBP1 and MAPK10 in the PoOxSmoke analysis of the European sample and TJP3 in the PoOxVitamin analysis of the pooled sample. The current study was primed to explore new hypotheses for disease mechanisms and to provide as many of the results as possible so that other researchers with access to similar GWAS datasets would be able to validate the findings presented here. To avoid being overly stringent, we thus presented all the results for the top 20 SNPs in Table 3–Table 5.
Despite an exhaustive literature search, we were unable to find any obvious evidence linking ANK3 and orofacial clefts. The Hetionet results confirmed this lack of a direct connection (Figure 1). ANK3 encodes a member of the Ankyrin family of proteins, whose function is to bind the integral membrane proteins to the spectrin-actin cytoskeleton. This is important for cell motility, activation, proliferation, contact and the maintenance of specialized membrane domains; cellular activities that are also relevant for the proper development of craniofacial structures. For example, Stankewich and colleagues54 showed that the spectrin–ankyrin scaffold is important for cell migration, tissue patterning and organogenesis. Homozygous deletion of the gene encoding αII-spectrin in mice (Spna2) resulted in craniofacial, neural tube and cardiac anomalies, in addition to retarded intrauterine growth. Figure 3 indicates that several SNPs in ANK3 are potentially associated with clefting.
Like ANK3, ARHGEF10 has not previously been associated with orofacial clefts, and the Hetionet results are consistent with this observation (Figure 2). ARHGEF10 encodes a Rho guanine nucleotide exchange factor that may be involved in neural morphogenesis55. LYZL1 belongs to the family of lysozyme-like proteins that are implicated in sperm function and innate immunity56. According to GeneCards (GCID: GC09P020659), FOCAD encodes a tumor suppressor gene that is highly expressed in the brain. It has also been linked to Alzheimer's disease57. Furthermore, germline deletions in FOCAD are associated with polyposis and colorectal cancer58. Again, as with ANK3 and ARHGEF10 above, there do not seem to be any obvious connections between LYZL1 or FOCAD with clefting.
In contrast to the above genes, FRAS1 and several members of the platelet-derived growth factor (PDGF) gene family are known to be implicated in orofacial clefts. PDGFD is a member of the PDGF gene family and plays a central role in the PDFG receptor-alpha (PDGFR-α) signaling pathway. More specifically, disruption of Pdgf signaling results in clefting of the palate59. FRAS1 (GCID: GC04P078056) encodes an extracellular matrix protein that plays a critical role in epithelial-mesenchymal interactions during embryonic development60. Loss-of-function mutations in FRAS1 underlie Fraser syndrome, which is characterized by craniofacial, urogenital and respiratory system abnormalities61. Both of these genes are therefore worthy of further investigations in other isolated orofacial cleft cohorts.
A limitation of this study is that genotypes were not imputed. To avoid Mendelian inconsistencies, the imputation procedure would have had to account for the full triads, as opposed to imputing each sample independently, which has not been done for our data. Instead we conducted post hoc haplotype analyses for combinations of top SNPs located close to each other. This is akin to imputation, in that such an analysis takes into account information about a whole area of DNA, instead of just one SNP. Table 7 shows that in the PoOxSmoke analysis of the two-SNP combination rs3793861-rs7087489 in ANK3, the p-value was slightly lower and the RRR slightly higher than in the corresponding analyses of each individual SNP. A similar pattern was observed in the PoOxAlcohol analyses of the rs2294035-rs4876274 haplotype in ARHGEF10. This indicates that the two-SNP combinations may be driving the effects observed with the individual SNPs.
The genes PDGFD, CSMD1 and RSUI detected here had previously showed up in a study focusing on identifing GxE effects in the same CL/P triads28. In that study, a possible GxVitamin effect was detected with PDGFD and RSUI, and a possible GxAlcohol effect was detected with CSMD1. In the current study, a PoOxSmoke effect was detected with PDGFD and PoOxVitamin effects were detected with RSUI and CSMD1. In other words, only RSUI had the same exposure (vitamin) across the studies. RSUI stands for ‘Ras suppressor protein 1’ and is localized to chromosome 10p13. Its protein product is found at cell–extracellular matrix adhesion sites and has been reported to be involved in supressing v-Ras transformation in the Ras signal transduction pathway62. CSMD1 stands for ‘CUB and Sushi multiple domains 1’ and is localized to chromosome 8p23.2 (GCID: GC08M002953). It is involved in tumor suppression, as it has frequently been found to be deleted in many types of cancers63,64. Again, there does not seem to be any obvious connections to clefting.
None of the top SNPs identified in our previous study focusing on PoOxE effects in CPO triads overlapped with SNPs identified in this study of CL/P27. This is consistent with the observation that CPO and CL/P are etiologically distinct, so that the lead SNPs may be subtype-specific and differ between the two conditions. However, we detected associations with the ‘cytochrome P450 family 4 subfamily F member 3’ gene (CYP4F3 on chr 19p13.12) in the previous CPO analyses, and with the ‘cytochrome P450 family 46 subfamily A member 1’ gene (CYP46A1 on chr 14q32.2) in the present study. These two genes are members of the cytochrome P450 superfamily of enzymes that are primarily found in liver cells and whose function is to catalyze many reactions involved in the biotransformation of xeno- and endobiotics, and the biosynthesis of cholesterol and lipids, among others65. It is therefore not surprising that these genes would appear in an analysis focusing on smoking, alcohol and vitamin intake.
We searched Hetionet for indirect links between ANK3 or ARHGEF10 and cleft lip, as well as between ANK3 or ARHGEF10 and nicotine or alcohol dependence, respectively. This approach has several limitations. First, using “nicotine dependence” and “alcohol dependence” in lieu of the actual smoking and alcohol consumption status may introduce some bias. Second, Hetionet is built from a curated set of database information, which means that not all the information, especially the newest, would be available. However, when interpreting our results, we used the source databases to make sure that the connections between the nodes are reliable.
To conclude, our search for an interaction between a PoO-effect and an environmental exposure for CL/P identified possible relationships between SNPs in ANK3 and maternal smoking, and SNPs in ARHGEF10 and maternal intake of alcohol. There is a possibility that these interactions have a biological basis, although without replication they remain speculative. Our demonstration of the feasibility of identifying complex interactions between relevant environmental exposures and PoO-effects opens new possibilities in the search for the genetic etiology of CL/P.
The GWAS data are available in the dbGaP database. Additional information regarding the inclusion/exclusion criteria of the study, the ethics statements, data variables, study history, publications, and other documentation related to the study is provided on the dbGaP website.
The dbGaP database at the National Center for Biotechnology Information, U.S. National Library of Science (NCBI/NLM) provides an extensive overview of the cleft dataset used in this study. Entering the dbGaP accession number phs000094.v1.p1 provides access to information regarding the variables, study documents, and datasets. For example, detailed information about the mother’s exposure to alcohol, vitamins, and smoke is provided under the header “Variable Selection”. Information on study questionnaires, institutional review boards and consent forms from each participating cohort can be found under the header “Documents”.
Controlled-access data from dbGaP is available only through the dbGaP authorized access portal. There are separate procedures for accessing individual data, depending on whether the researcher is NIH-affiliated (intramural) or not (extramural). In addition, other restrictions apply; e.g., the principal investigator from the applying institution needs to be a permanently employed professor, senior scientist, or equivalent, to submit a data access request. A valid eRA Commons account for logging in to the dbGaP system is also mandatory.
- Source code available from: https://github.com/oeh041/A-genome-wide-scan-of-cleft-lip-triads-identifies-parent-of-origin-interaction-effects-between-ANK3-
- Archived source code at time of publication: https://doi.org/10.5281/zenodo.324131938
- License: CC-BY 4.0
This research was supported by the Bergen Medical Research Foundation [807191], by the Research Council of Norway (RCN) through its Centres of Excellence funding scheme [grant 262700], and by the Biobank Norway II from the RCN [245464].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We extend our warmest gratitude to the families from so many different countries in the world who opted to contribute to this study. We are also grateful to everybody who contributed to the collection of the data, from recruiters in the field to technicians in the lab.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genetic epidemiology of childhood disorders
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genetics of craniofacial diseases
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genetic epidemiology of childhood disorders
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 19 Jul 19 |
read | |
|
Version 1 24 Jun 19 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)