Keywords
Olsenella, culturomics, chicken gut microbiota, taxono-genomics
Olsenella, culturomics, chicken gut microbiota, taxono-genomics
In this version, the manuscript was revised according to the reviewers’ comments. All suggestions have been addressed. Some grammar corrections were performed. The tittle of the Table 1 was replaced with “Phenotypic characteristics”. The species names were used instead of numbers. The Bioproject number was removed and GenBank SRA accession number was applied in the Table 3.
See the authors' detailed response to the review by Yung-Fu Chang
The chicken gut harbors highly diverse microbes1. The gut microbes are known for their nutritional benefits by producing short-chain fatty acids, enzymes, amino acids along with their ability to resist pathogens, immunity development and maintain homeostasis2. Even though culture-independent methods have highlighted the functional capability of gut microbes, validation of these functions requires their cultivation, identification, and characterization. Most of the intestinal bacteria have been never isolated in the laboratory3,4 thus hindering the understanding of their ecological and functional roles in the gut. Recently, “culturomics” strategy drives the discovery of previously uncultured species based on modified culture conditions, such as media, temperature, pH and atmosphere and rapid identifying methods; matrix-assisted desorption ionization- time of flight mass spectrometry (MALDI-TOF MS) and 16S rRNA gene sequencing5–7. We employed culturomics to isolate bacteria from the cecum of feral chickens. Based on bacterial isolation and identification, strain SW165T was found as a new species within the genus Olsenella.
The members of the genus Olsenella are strictly anaerobic, Gram-positive, non-motile, non-spore-forming bacilli, or cocci. This genus was first named by Dewhirst et al. 20018 and amended by Zhi et al 20099 and Kraatz et al 201110. This genus has recently been reclassified as a member of the family Atopobiaceae under order Coriobacteriales, class Coriobacteria, and phylum Actinobacteria11. The genus Olsenella consists of nine published species; O. uli12, O. profusa8, O. umbonata10, O. scatoligenes13, O. urininfantis14, O. congonensis15, O. provencensis16, O. phocaeensis16, and O. mediterranea16. The members of this genus are strictly anaerobic, Gram positive, non-motile, non-spore forming rod-shaped with G+C content of DNA 62–64%8,13,17. The main habitats of the Olsenella are the oral cavity and gastrointestinal tract of humans18–21, animals and various anaerobic environmental sites22–24. In the chicken cecum, many members of the genus Olsenella have been reported in the chicken microbiome in metagenomic-based studies15,25. However, only O. uli was isolated from chicken gut26.
The taxono-genomics approach uses a combination of phenotypic and genotypic characterization to describe new bacteria27,28. Phenotypic investigation includes morphological, physiological, and biochemical assays. Genome-based and 16S-based analyses are used in genotypic characterization. In this study, strain SW165T was described using taxono-genomics and compared to its closely related phylogenetic neighbors. Following analysis, we found that strain SW165T belongs to a novel species for which the name Olsenella lakotia SW165T sp. nov. is proposed.
The cecal content of feral chickens was collected in Brookings, South Dakota, USA. For cultivation, the samples were transferred into an anaerobic workstation (Coy Laboratory) containing 85% nitrogen, 10% hydrogen and 5 % carbon dioxide and plated on a modified Brain Heart Infusion (BHI-M) agar containing 37 g/L of BHI, 5 g/L of yeast extract, 1 ml of 1 mg/ml menadione, 0.3 g of L-cysteine, 1 ml of 0.25 mg/L of resazurin, 1 ml of 0.5 mg/ml hemin, 10 ml of vitamin and mineral mixture, 1.7 ml of 30 mM acetic acid, 2 ml of 8 mM propionic acid, 2 ml of 4 mM butyric acid, 100 µl of 1 mM isovaleric acid, and 1% of pectin and inulin. After 3 days of incubation at 37°C under anaerobic conditions, a single colony of strain SW165 was identified by MALDI-TOF mass spectrometry using a Microflex spectrometer (Bruker Daltonics, Bremen, Germany). The strain was maintained in BHI-M medium and stored with 10% (v/v) Dimethyl Sulfoxide (DMSO) at -80°C.
For morphological characterization, the strain SW165T was anaerobically cultivated in BHI-M medium, pH 6.8-7.2, at 37o C. Colony morphologies were examined after 2–3 days of incubation on BHI-M agar plates. Gram-staining was performed using a Gram-Staining kit set (Difco), according to the manufacturer’s instructions. Cell morphologies were examined by scanning electron microscopy (SEM) of cultures during exponential growth. Aerotolerance was examined by incubating cultures for 2 days separately under aerobic and anaerobic conditions. Growth of strain SW165T at 4, 20, 30, 37, 40 and 55°C was determined. For determining the range of pH for growth of SW165T, the pH of the medium was adjusted to pH 4.0–9.0 with sterile anaerobic stock solutions of 0.1 M HCl and 0.1 M NaOH. The motility of this microorganism was determined using motility medium with triphenyltetrazolium chloride (TTC)29. The growth was indicated by the presence of red color, reduced form of TTC after it is absorbed into the bacterial cell wall.
Biochemical tests to determine standard taxonomic characteristics for strain SW165T were performed in triplicate. The utilization of various substrates as sole carbon and energy sources and enzyme activities were performed using the AN MicroPlate (BIOLOG) and API ZYM (bioMérieux) according to the manufacturer's instructions. Reference strain, DSM 13989T purchased from the DSMZ culture collection and isolated strains SW165T were simultaneously cultured in BHI-M medium at 37°C for 24 h under anaerobic condition before cell biomass were harvested for cellular fatty acid analysis. The fatty acids were extracted, purified, methylated, and analyzed using gas chromatography (Agilent 7890A) based on the instruction from Microbial Identification System (MIDI)30. Metabolic end-products such as short-chain fatty acids of strain SW165T and DSM 13989T grown in BHI-M were determined using a gas chromatography. The cultures were deproteinized with 25% metaphosphoric acid before supernatant collection. The supernatant was analyzed for the presence of acetic acid, butyric acid, isovaleric acid, and propionic acid using GC (Themo ScientificTM TRACETM 1310 GC equipped with a TraceGOLDTM TG-WaxMS A GC column.).
Genomic DNA of the strain SW165T was extracted using a DNeasy Blood & Tissue kit (Qiagen), according to the manufacturer’s instructions. 16S rRNA gene sequence was amplified using universal primer set 27F (5’- AGAGTTTGATCMTGGCTCAG-3’; Lane et al., 1991) and 1492R (5’- ACCTTGTTACGACTT- 3’; Stackebrandt et al., 1993)31,32, and sequenced using a Sanger DNA sequencer (ABI 3730XL; Applied Biosystems). The 16S rRNA gene sequence of SW165 was then compared with closely related strains from the GenBank (www.ncbi.nlm.nih.gov/genbank/) and EZBioCloud databases (www.ezbiocloud.net/eztaxon)33. Alignment and phylogenetic analysis were conducted using MEGA7 software34. Multiple sequence alignments were generated using the CLUSTAL-W35. Reconstruction of phylogenetic trees was carried out using the maximum-likelihood (ML)36, maximum-parsimony (MP)37, and neighbor-joining (NJ)38 methods. The distance matrices were generated according to Kimura's two-parameter model. Bootstrap resampling analysis of 1000 replicates was performed to estimate the confidence of tree topologies.
The whole genome sequencing of strain SW165T was performed using Illumina MiSeq sequencer using 2x 300 paired end V3 chemistry. The reads were assembled using Unicycler that builds an initial assembly graph from short reads using the de novo assembler SPAdes 3.11.139. The quality assessment for the assemblies was performed using QUAST5.0.240. Genome annotation was performed using Rapid Annotation using Subsystem Technology (RAST) server41. The digital DNA-DNA hybridization (dDDH) was performed using Genome-to-Genome Distance Calculator (GGDC) web server (http://ggdc.dsmz.de) to estimate the genomic similarity between strain SW165T and the closest phylogenetic neighbor. Average nucleotide identity (ANI) between strain SW165T and the closely related strains was also calculated using the OrthoANI software42. Distribution of functional categories of strain SW165T was compared to Olsenella species and was presented in a heatmap generated using Explicet version 2.10.543.
Strain SW165T was isolated from cecal contents of feral chicken in an anaerobic chamber (Coy Laboratory Product, MI, USA). Colonies of SW165T on BHI-M agar were 0.2–0.5 cm in diameter, appeared white, smooth, and umbonate with entire circular edges when grown at 37°C anaerobically after 48 hours of incubation. After cultivation, the colonies of this strain were subjected to identification by MALDI-TOF using a Microflex spectrometry (Bruker Daltonics, Bremen, Germany). MALDI-TOF did not identify the strain as the scores obtained were < 1.70. Thus, full length 16S rRNA gene was sequenced using Sanger sequencing method. The 16S rRNA of the strain SW165T showed 96.33% identity with O. profusa DSM 13989T (GenBank accession no. AF292374), the validly closest species within phylogenetical nomenclature (Figure 1). The current cut off for species delineation from its nearest neighbor based on 16S rRNA is 98.7%44. As the identity of 16S rRNA of strain SW165T was lower than threshold, it was considered as a representative of putatively novel species within the genus Olsenella in the family Atopobiaceae. Phylogenetically, the strain was found to cluster together with other members of genus Olsenella, as shown in Figure 1, validating that SW165T belongs to genus Olsenella taxonomically.
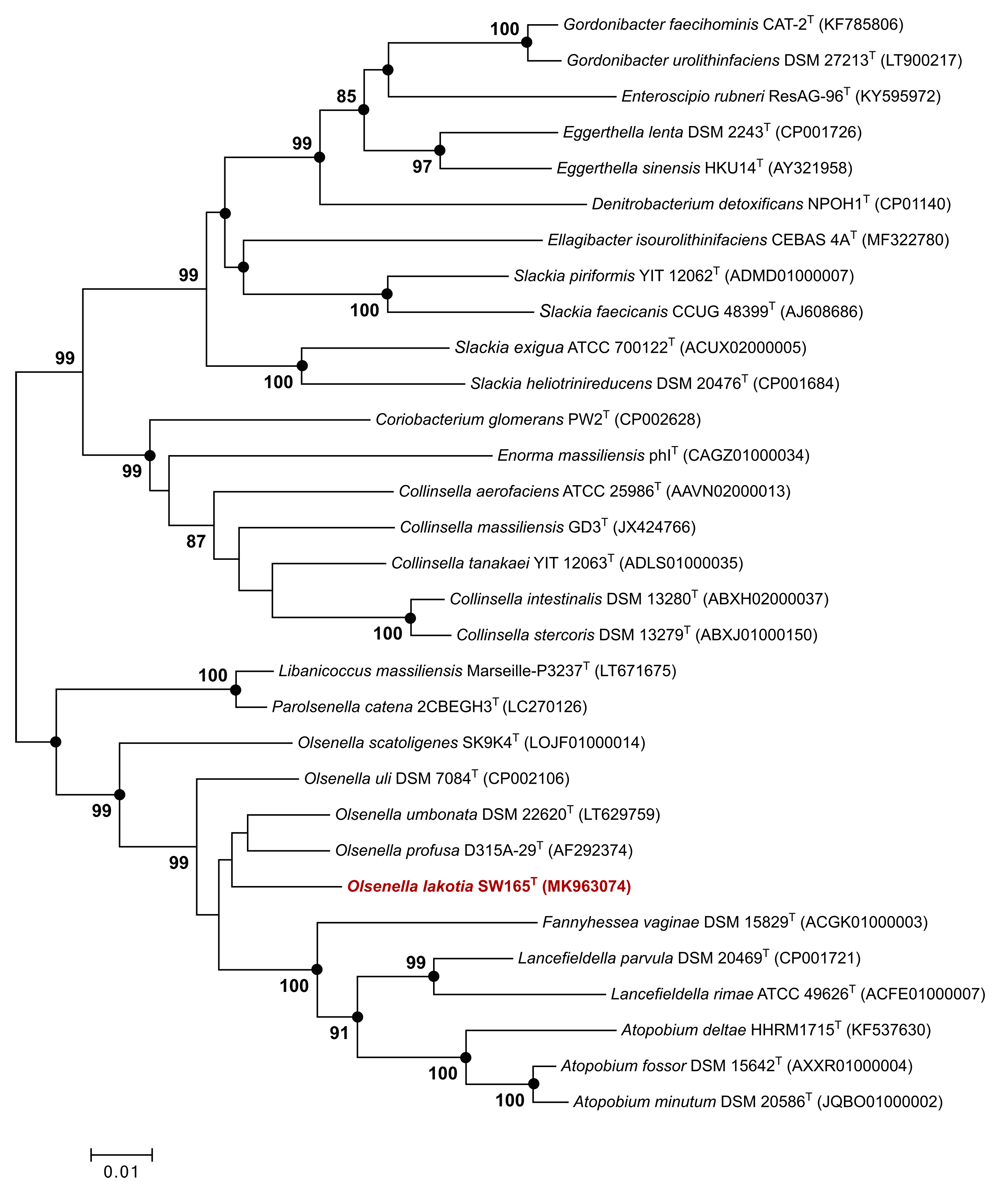
Tree shows phylogenetic position of Olsenella lakotia DSM 107283T and closely related species in the family Atopobiaceae. GenBank accession numbers of the 16S rRNA gene sequences are given in parentheses. Black circles indicate that the corresponding branches were also recovered both by maximum-likelihood and maximum parsimony methods. Bootstrap values (based on 1000 replications) greater than or equal to 70% are shown in percentages at each node. Bar, 0.01 substitutions per nucleotide position.
Phenotypic growth of strain SW165T was observed on modified BHI-M agar after 2–3 days of incubation at temperature between 37°C and 45°C and pH between 6.0–7.0. The optimum temperature and pH for the growth were at 45°C and pH 7.0, respectively. Strain SW165T grew only under anaerobic conditions, suggesting obligate anaerobic nature. Bacterial cells were Gram-stain-positive bacilli (0.5–2.0 µm), growing in pairs or as short chains, and were non-motile (Figure 2).
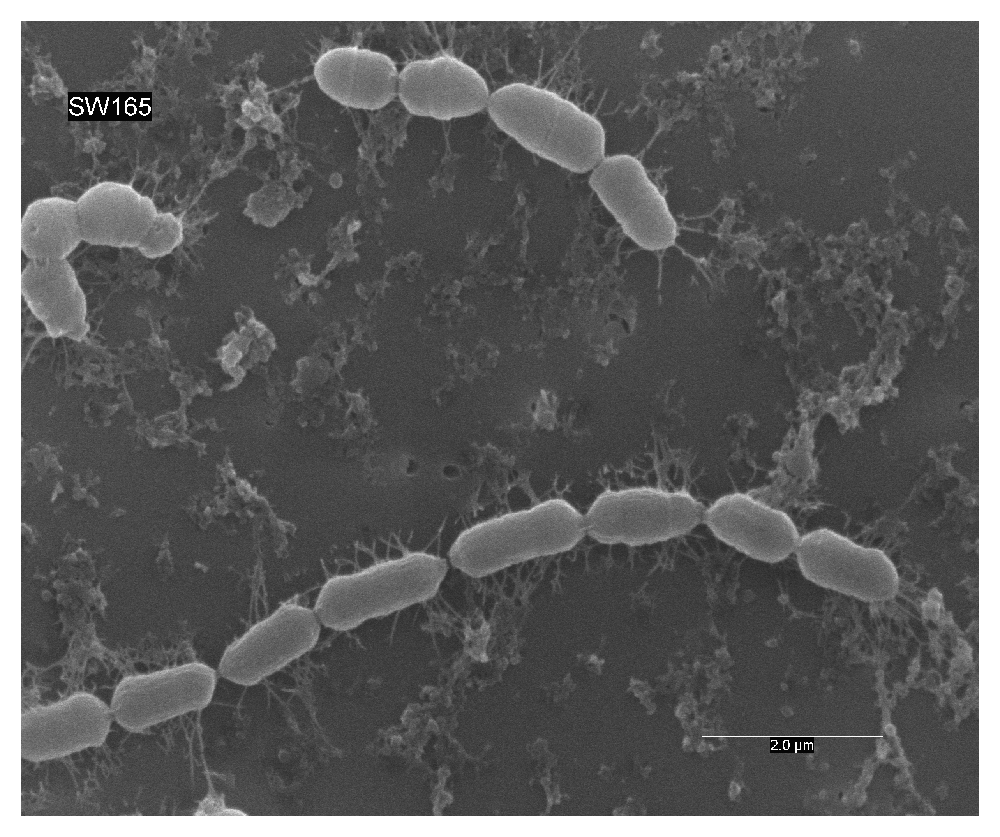
Cells were anaerobically cultured for 24 hours at 37°C in BHI-M medium. Bar, 2 μm; uncropped/unedited image.
To further analyze the biochemical properties of the strain, we performed the carbon source utilization assay using BIOLOG AN microplate and compared it to closely related taxa. Strain SW165T consumed various carbon sources for the growth, which differed from related strains in the utilization of D-fructose, L-fucose, D-galactose, maltose, D-melibiose, and D-raffinose, and in the non-utilization of dulcitol. Based on the enzymatic activity test, the strain produced several enzymes, including alkaline phosphatase, leucine arylamidase, cysteine arylamidase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, and β-glucosidase. Interestingly, alkaline phosphatase α-galactosidase, β-galactosidase and β-glucuronidase are not reported from its closest neighbors (Table 1). Furthermore, the dominant cellular fatty acids of the strain SW165T were saturated, including C 12 : 0 (25.5%) and C 14 : 0 (22.83%). Moreover, other dominant fatty acids were C 14 : 0 DMA (15.61%) and summed feature 1 [C13:1 and/or C14:0 aldehyde; 13.94%]. However, there were distinct quantities of some fatty acids between SW165T and the relative strains (Table 2). The major short chain fatty acids produced by SW165 when cultivated in BHI-M were acetic acid (3.74 mM) followed by propionic acid (0.53 mM).
Column headers show Strains designated: SW165T; O. profusa DSM 13989T; O. uli DSM 7084T; O. umbonata DSM 22620T; O. scatoligenes DSM 28304T. Results for metabolic end products of SW165 are from this study with cells that were cultured for 3 days at 37°C in BHI-M. +, positive; -, negative; w, weak; ND, not determined.
| Characteristic | O. lakotia SW165T | O. profusa DSM 13989T | O. uli DSM 7084T † | O. umbonata DSM 22620T † | O. scatoligenes DSM 28304T ‡ |
|---|---|---|---|---|---|
| Gram stain | + | + | + | + | + |
| Growth at 37° C | + | + | + | + | + |
| Motility | - | - | - | - | - |
| Carbon source (BIOLOG AN) | |||||
| Arbutin | + | + | ND | ND | ND |
| D-Cellobiose | + | + | - | - | + |
| Dextrin | + | + | ND | ND | ND |
| D-Fructose | + | - | ND | ND | ND |
| L-Fucose | + | - | ND | ND | ND |
| D-Galactose | + | - | ND | ND | ND |
| α-D-Glucose | + | + | ND | ND | ND |
| Dulcitol | - | + | ND | ND | ND |
| Maltose | + | - | - | + | ND |
| D-Mannose | + | + | - | + | ND |
| D-Melibiose | + | - | - | - | ND |
| D-Raffinose | + | - | ND | ND | ND |
| Salicin | + | + | - | - | + |
| Sucrose | + | + | - | + | ND |
| Turanose | + | + | ND | ND | ND |
| Enzyme activity (API ZYM) | |||||
| Alkaline phosphatase | + | - | + | - | ND |
| Esterase (C 4) | - | - | + | + | - |
| Leucine arylamidase | + | + | + | + | ND |
| Cystine arylamidase | + | + | - | - | ND |
| α-chymotrypsin | w | - | ND | ND | ND |
| α-galactosidase | + | - | ND | ND | ND |
| β-galactosidase | + | - | - | - | + |
| β-glucuronidase | + | - | ND | ND | ND |
| α-glucosidase | + | + | - | + | + |
| β-glucosidase | + | + | + | - | + |
| Short-chain fatty acid production | A | L, a, f | L, a, f | L, a, f | L, a, f |
| DNA G+C content (mol%) | 67.59 | 64 | 64.7 | 64.9 | 62.1 |
Values are percentages of total fatty acids detected. Fatty acids with contents of less than 1% in all strains are not shown; ND, Not detected.
| Fatty acid composition | O. lakotia SW165T | O. profusa DSM 13989T | O. uli DSM 7084T ‡ | O. umbonate DSM 22620T ‡ | O. scatoligenes DSM 28304T ‡ |
|---|---|---|---|---|---|
| Straight chain | |||||
| C10 : 00 | 3.34 | ND | ND | ND | ND |
| C12 : 00 | 25.5 | ND | 2.8 | ND | ND |
| C14 : 00 | 22.83 | 9.94 | 1.3 | 31.6 | 25.9 |
| C16 : 00 | 2.69 | 5.82 | 4.3 | 6.2 | 2.7 |
| C16 : 0 aldehyde | 0.92 | 3.48 | ND | ND | ND |
| Demethylacetal (DMA) | |||||
| C12 : 0 DMA | 8.44 | ND | ND | ND | ND |
| C14 : 0 DMA | 15.61 | 4.33 | ND | ND | ND |
| C16 : 0 DMA | 1.21 | 13.97 | ND | ND | ND |
| C18 : 0 DMA | 0.65 | 1.18 | ND | ND | ND |
| Branched | |||||
| C14 : 0 iso | ND | 22.34 | ND | ND | ND |
| C13 : 0 anteiso | ND | 1.79 | ND | ND | ND |
| C15 : 0 anteiso | ND | 14.48 | ND | ND | ND |
| C15 : 0 anteiso DMA | ND | 5.17 | ND | ND | ND |
| Unsaturated | |||||
| C18 : 1ω9c | 1.82 | 3.9 | 69.8 | 20.7 | 25.7 |
| C18 : 2ω6,9c | 0.89 | 2.03 | ND | ND | ND |
| Summed Feature 1 | 13.94 | 2.15 | ND | 11.3 | 20.7 |
| Summed Feature 13 | ND | 5.17 | ND | ND | ND |
‡Data from et al. (2015)13
*Summed features are fatty acids that could not be separated using the MIDI System. Summed feature 1 contains C13 : 1 and/or C14 : 0 aldehyde. Summed feature 13 contains C15 : 0 anteiso and/or C14 : 0 2-OH.
We examined the genome of the strain SW165T to investigate its differentiation from the neighbors. The genome size of strain SW165T was 2,427,227 bp with 67.59 mol% G+C content. The draft genome was assembled into 33 contigs with 2,228 protein-coding sequences and 52 RNAs (Table 3) and is visualized as Figure 3. The genomes sizes for the Olsenella species were comparable to one another except for O. urininfantis whose was only 1.75 Mbps. However, the G+C contents strain SW165T and O. mediterranea were the highest but comparable to other neighboring Olsenella species (Table 3). The genome of SW165T possessed a total of 1, 230 genes with putative function and 998 genes as hypothetical proteins. Among 1,230 genes, 823 were classified as features in subsystem, following functional categories (COGs). The majority of categories included amino acid and derivatives (172 genes), carbohydrates (163 genes), and protein metabolism (132 genes) (Extended data: Supplemental Table 145).
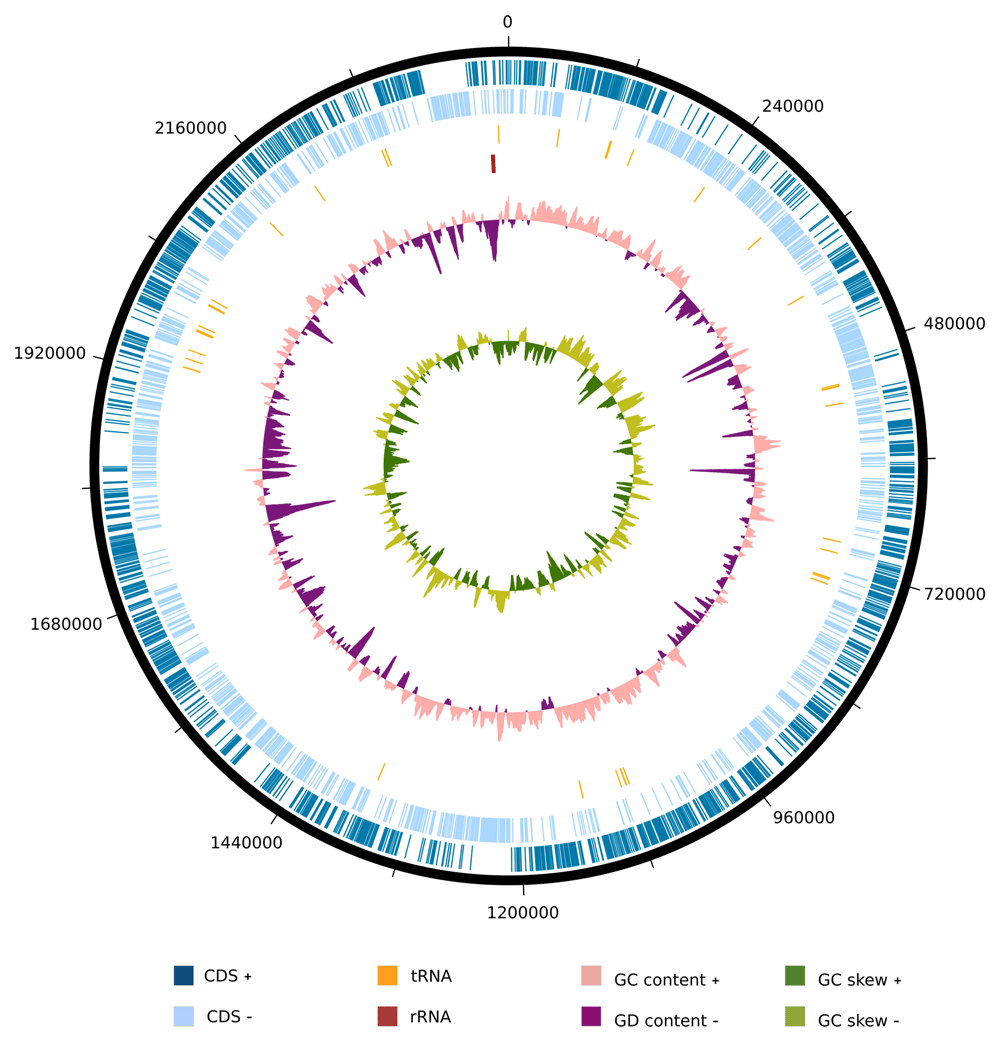
From outside to the center: coding sequences on the forward strand (CDS +), coding sequences on the reverse strand (CDS -), tRNAs, rRNAs, GC content, and GC skew.
Furthermore, we compared the genome of SW165T to its neighbor using OrthoANI as shown in Figure 4. The genome of SW165 was only 73.41% identical to its nearest neighbor O. profusa DSM 13989T. Also, the OrthoANI values for SW165T and closely related strains ranged from 65.40 to 74.18 % (Figure 4) indicating that the genome of SW165T is unique compared to its neighbors. The proposed cut off for OrthoANI for the species delineation is 95–96% identity46,47. In addition, dDDH between SW165T and the closest neighbor, O. profusa DSM 13989T was only 17.6 ± 5.3. These values were lower than threshold of ANI and dDDH for delineating prokaryotic species, suggesting that these strains are distinct species. Also, the gene distribution into COGs was comparable in all eight compared Olsenella genomes (Figure 5). Hence, the phenotypic and genetic discrepancy of the SW165T with its close neighbor apparently supports that strain SW165T represents a new species of the genus Olsenella.
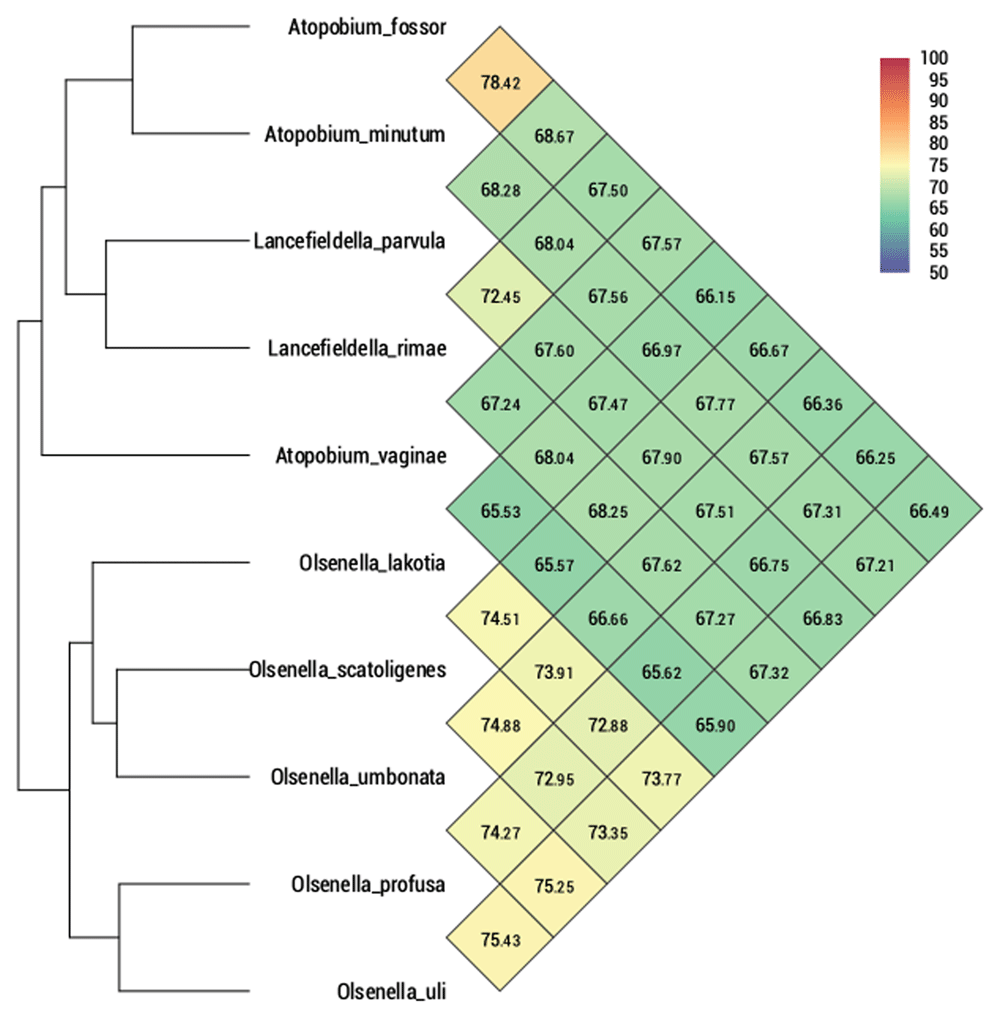
Heatmap represents OrthoANI values generated using OAT software between O. lakotia and related taxa with valid taxonomy.
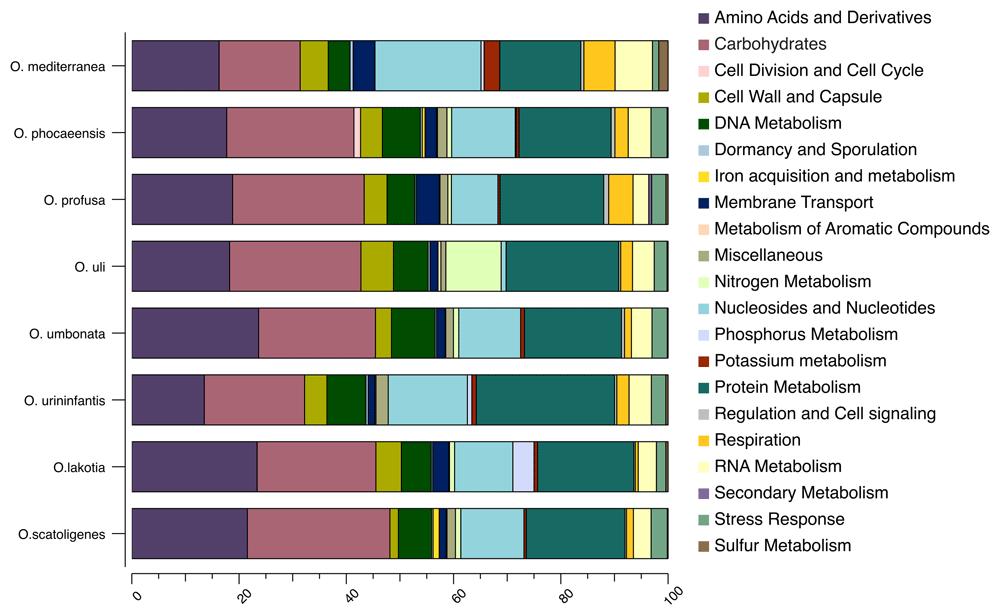
The functional features were predicted based on the clusters of orthologous groups. Heatmap was generated from the genome annotation of individual species by RAST using Explicet software.
Culturomics of the gut microbiota has evolved as a strong tool to increase the isolation of diverse previously uncultured bacteria from the gut5,48. The cultivation of the gut microbiota enables to improve the health through an enhanced understanding of their roles in the gut ecosystem and finally to the host. Thus, using the culturomics strategy, we were able to isolate previously uncultured bacterium SW165T from cecal content of feral chicken and finally characterize and describe it using taxono-genomics as a novel microorganism.
The novelty of a prokaryotic organism is universally determined by the comparison of 16S rRNA gene sequence homology49. The threshold values are used at distinct taxonomic levels46. In this context, the newly discovered bacterium was initially validated using the full-length 16S rRNA gene sequences, which were thereafter used for taxonomic classification. Phylogenetic analysis of 16S rRNA gene showed that the novel strain SW165T clustered with closely related taxa in the genus Olsenella within the family Atopobiaceae (Figure 1). This genus composes of nine species, most of which are members of the gut microbiota of humans and animals. However, O. uli is only a species that have been isolated from chicken gut26. Remarkable, this study revealed a new member of Olsenella from gut microbiota of chicken.
Phenotypic analyses are performed to differentiate closely valid bacteria. Based on phenotypic tests, strains SW165T appeared several distinct properties compared to other members of the genus Olsenella (Table 1 and Table 2). The obvious distinctive phenotypic features were observed in biochemical tests including enzymatic activity and carbon source utilization, thereby they might be important parameters for discriminating closely related species. These differences suggested the novelty of this microorganism belonging to Olsenella.
In addition to 16S based comparison, whole genome can be used for distinguishing, different bacteria. Recently, digital DNA-DNA hybridizations (dDDH) becomes a key measurement in the delineation of prokaryotic species. It is an in-silico genome-to-genome comparison inferring whole genome distance to mimic DDH50. Besides, Average Nucleotides Identification (ANI) is another particular tool that confirms the taxonomic delineation. It measures the overall similarity between two genome sequences51. A recent publication of novel bacteria trend to perform genome-based analysis to support the results of 16S rRNA gene-based analysis. The strengths of genome-based analyses include a comparison of all nucleotides in prokaryotic taxonomy and functional prediction52. Based on genomic evidence, strain SW165T showed low similarity in terms of OrthoANI with Olsenella species of the family Atopobiaceae (Figure 4). Further, genome features and distribution of predicted functional categories of strain SW165T has corresponded to all other Olsenella species (Figure 5 and Table 3). Thus, we proposed the strain SW165T as a new species Olsenella lakotia SW165T sp. nov., within the family Atopobiaceae.
O. lakotia sp. nov. (la.ko’tia N.L n. referring to native American tribe). Cells are strictly anaerobic, Gram-positive streptobacillus and non-motile. The average size of each cell is 0.5–2.0 µm. Colonies are visible on BHI-M agar after 2 days and are approximately 0.2–0.5 cm in diameter, cream-white, smooth, slightly umbonate with an entire circular margin. The microorganism exhibits optimal growth in the BHI-M medium at 45°C and pH 7.0. The strain utilizes arbutin, cellobiose, dextrin, D-fructose, L-fucose, D-galactose, α-D-glucose, maltose, D-mannose, D-melibiose, D-raffinose, salicin, sucrose, and turanose as a carbon source. Positive enzymatic reactions are obtained for alkaline phosphatase, leucine arylamidase, cysteine arylamidase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, and β-glucosidase. The volatile fatty acid produced by this strain is acetic acid. The primary cellular fatty acids are C12 : 0, C14 : 0, C14 : 0 DMA and summed feature 1. The genome is 2,427,227 bp, and its G+C is 67.59 mol%.
The type strain SW165T (=DSM 107283 =CCOS 1887) was isolated from the cecum of feral chicken was deposited in the DSMZ and CCOS collections under accession numbers DSM 107283 and CCOS 1887 (Extended data: Supplemental Data 153), respectively. The 16S rRNA and genome sequence are available in GenBank under accession numbers MK963074 and BioProject PRJNA545153, respectively.
Olsenella sp. strain SW165 16S ribosomal RNA gene, partial sequence, Accession number MK963074: https://www.ncbi.nlm.nih.gov/nuccore/MK963074
Olsenella lakotia SW165 Genome sequencing and assembly, Accession number PRJNA545153: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA545153
Figshare: Extended data; Supplemental Table 1 (Functional categories (COGs) from genome of strain SW165T), https://doi.org/10.6084/m9.figshare.12793544.v145.
Figshare: Supplemental Data 1 (DSMZ and CCOS accession numbers of strain SW165T), https://doi.org/10.6084/m9.figshare.12793610.v153.
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
SW gratefully acknowledges Science Achievement Scholarship of Thailand (SAST) for providing fellowship. The authors would like to thank Electron Microscopy Core Facility at the Bowling Green State University, Ohio, USA for assistance with scanning electron microscopy.
A previous version of this article was published on bioRxiv: https://doi.org/10.1101/670927
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Microbiology
Is the work clearly and accurately presented and does it cite the current literature?
No
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: microbial pathogenesis, molecular diagnosis, vaccine development, genomics, and proteomics.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 08 Oct 20 |
||
|
Version 1 08 Sep 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)