Keywords
CPRD, Primary Care, Electronic Healthcare Records, Epidemiology, R
This article is included in the RPackage gateway.
CPRD, Primary Care, Electronic Healthcare Records, Epidemiology, R
The Clinical Practice Research Datalink (CPRD) is an NHS primary care service that stores clinical, referral, therapeutic data and linked medical services such as imaging and hospital records of patients enrolled in England (UK). In its current form, CPRD has been active for over twenty years and holds up to 11 million records1. CPRD data has been widely used to improve medical practice and to further our understanding of drug efficacy and drug safety and disease mechanism2. As with all longitudinal data, understanding the outcome of a disease is usually confounded by several factors3. Patient comorbidities, therapy data and social habits (to name but a few) are all potential confounders that should be treated appropriately (see 4 for a research example of common patient characteristics using a CPRD data release). However, the curation and manipulation of CPRD data can cause significant barriers for many researchers, especially those less familiar with manipulating text-based large datasets or programming.
Data released from the CPRD Gold database data is presented as a series of flat files. A patient’s longitudinal data is presented across several rows, with each column denoting a record property. Patient records are linked between flat files using an anonymized patient identifier and several additional identifiers depending on the data at hand. Clinical diagnosis and prescribed therapeutics is encoded as a medcode and a prodcode, with a corresponding description available using the CPRD data dictionary. Most requests for CPRD Gold primary care data will usually return several clinical flat files. These are primary care records added by the patient’s General Practitioner (GP). The GP may also record additional clinical information, such as whether the patient smokes, how much they weigh, how often they drink, or the results from medical tests. This data is contained in a linked additional clinical set of flat files.
We present an R script that we have used to retrieve additional clinical data for patients. The script contains the code necessary to retrieve several patient characteristics (smoking, alcohol consumption, etc.) and is relatively straight forward to extend. We aim to continue updating our GitHub release with further additional clinical properties and any corresponding medcode descriptions whilst our projects remain active. In this manuscript, we outline the link between clinical and additional clinical patient data, the execution flow of the R scripts, and how to expand on the existing code. The R script is available for free and to download under an MIT license on GitHub: https://github.com/acnash/CPRD_Additional_Clinical
We describe the link between patient clinical data and additional clinical data in cases where such a CPRD data release has been made available.
Both clinical data and additional clinical data are stored in two separate sets of files, often denoted as head_Extract_Clinical_##.txt and head_Extract_Additional_###.txt, respectively (this may depend on the service provided by those who extract the CPRD data). Additional clinical data, stored in the second set of files, is typically accompanied by a lookup data table with lookup codes stored in text files. Additional clinical data is identified by a unique entity (enttype, the entity type) value, for example, smoking status has an entity value of 4. Both clinical files and additional clinical files will have an enttype column. A researcher can retrieve all patients with a smoking status using this column. Then, depending on the particular additional information of interest, data on that entity can be retrieved from up to seven data fields, for example, whether they smoke, how many packs of cigarettes per week, whether they smoke a pipe, and how many ounces of tobacco. Some values are encoded and require the corresponding lookup files which share the same names as the entry in the data lookup column. For example, smoking status is defined in the first data column and the values are found in the YND.txt lookup file as denoted in the corresponding first data lookup column.
We have built an extensible R script which parses the clinical and additional clinical text files for an entity and then returns all clinical data with the corresponding additional clinical data for those patients with an entity of interest on record. As this is longitudinal data, patients may have several additional clinical rows on record for one entity. Therefore, the adid value (present in both sets of records) is used to match the clinical data with additional clinical data. If there are additional clinical records without a matching clinical record, the code ignores and moves to the next patient. We have also added medcode descriptions for those entities we are currently using. As our research expands so too will this list.
The R code is easy to expand and allows the researcher to treat each entity according to their needs. As the code stands, it currently supports our active research and we have been able to retrieve the additional clinical values and paired clinical medcode descriptions for smoking, alcohol consumption, exercise, and weight/BMI. A user need only execute several short lines of R code to retrieve any of the entities. For example, to retrieve patient data on smoking habits:
#source the R core
source("CPRDLookups.R")
#define the location of both sets of files
additionalFiles <- "./head_Extract_Additional_folder"
clinicalFiles <- "./Clinical_folder"
#add the file locations to a list with element names
additionalFileList <- list(additional=additionalFiles, clinical=clinicalFiles)
#look up smoking additional data
resultDF <- getEntityValue("smoking", additionalFileList, idList)The idList is an R list or vector of patids (patient identifiers) of those patients of interest. The additionalFileList list contains the dedicated location of both sets of files. The content of these files is loaded into data frames and stored in the R environment. Ensure that there is enough system RAM available to open CPRD data files.
The entity value smoking is used to instruct the algorithm to look for all clinical and additional clinical data with an entity value of 4. A list of currently supported entities can be retrieved by executing getEntityValue(). All matching clinical data with additional clinical data (matched using the adid identifier if there are multiple rows per patient) are returned as a data frame with an additional column for medcode description. The medcode descriptions have been added into the R script and are used by the entity values currently supported.
The user executes one function and the existing R framework steps through a series of predefined functions before entering the user’s bespoke function (Figure 1). To extend the code to support additional clinical data, the researcher need only expand the if() statement in the function getEntityValue() with a function call to their own get<enttype>Data() function, following the format presented in the functions getExerciseData(), getSmokingData(), etc. Whether the researcher wishes to expand the results with a description of the medcodes is controlled by calling the defined function addMedcodeDescription(resultDF, <medcode_description_list>) within the if() statement. Both areas are illustrated below, with the potential code extension commented and a user defined entity name denoted with the place holder new_entity:
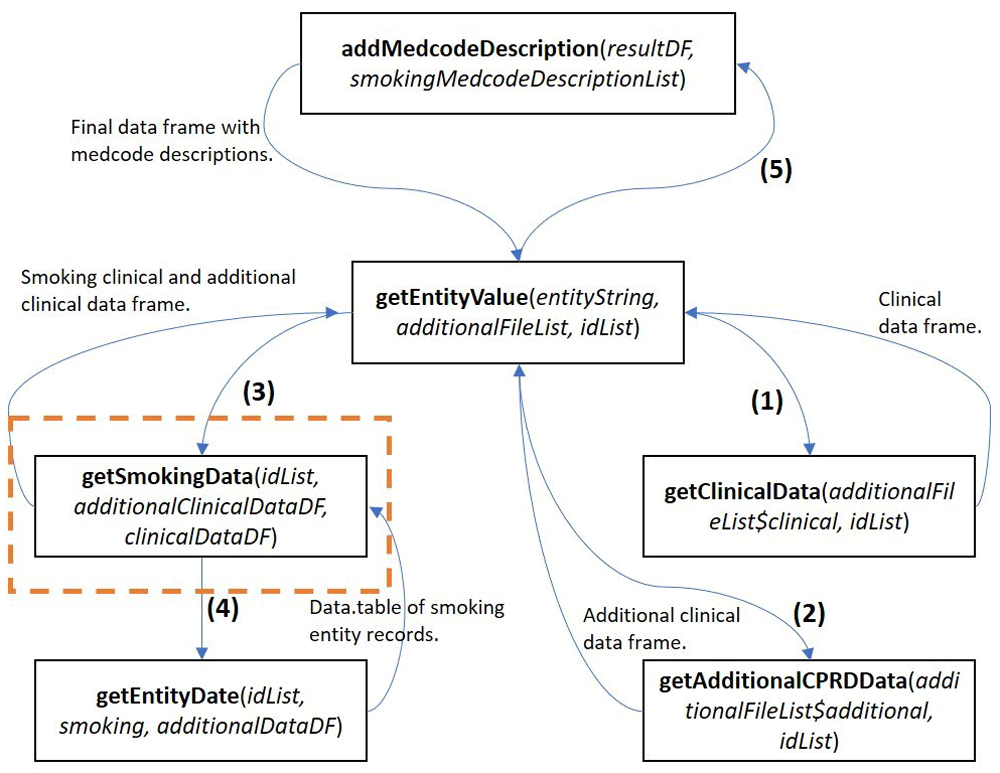
The function getSmokingData() in the orange square can be replaced by a user defined function with the same function arguments for other entities.
if(tolower(entityString) == "smoking") {
resultDF <- getSmokingData(idList, additionalClinicalDataDF, clinicalDataDF)
resultDF <- addMedcodeDescription(resultDF, smokingMedcodeDescriptionList)
} else if(tolower(entityString) == "bmi") {
resultDF <- getBMIData(idList, additionalClinicalDataDF, clinicalDataDF)
resultDF <- addMedcodeDescription(resultDF, bmiMedcodeDescriptionList)
}
#######
##Add more entity statements here....
#if(tolower(entityString) == “new_entity”) {
# resultDF <- getnew_entityData(idList, additionalClinicalDataDF, clinicalDataDF)
# resultDF <- addMedcodeDescription(resultDF, new_entityMedcodeDescriptionList)
#}
#######
else {
print("Unrecognised entity type. Try one of the following:")
outputCurrentOutput()
return(NULL)
}Unfortunately, it is not entirely possible to automate the decoding of the additional clinical data for every entity. There currently 501 unique entities (CPRD Gold release February 2018) and 91 unique lookup text files. As presented, we have used this code to return the decoded entity data along with a corresponding clinical medcode description. However, depending on the research, a user can return a result bespoke to their needs by simply defining a new function. Up to that stage, the existing code finds and retrieves all data with a matching enttype value.
The script requires the R data.table package.
We present a sample of a use-case where clinical records were linked with smoking patient data. The data presented in this manuscript and uploaded to the GitHub repository has been fabricated in the interest of patient confidentiality. A test data set of 2,000 clinical primary care records were parsed for additional data concerning smoking. Of those patients, 1,993 had a record for smoking. Having matched those patients, retrieved the data and decoded their smoking status for either “Data not entered”, “Yes”, “No” or “Ex smoker”, each medcode from a corresponding clinical record was populated with a readable description. A snapshot of fabricated patient records is presented in Figure 2.
We present an R script that searches for and retrieves additional clinical entity values and combines the results with patient clinical data. Although access to CPRD data requires an entrusted license holder, not all institutes will be fortunate enough to have access to software necessary to perform CPRD data curation and analysis5. At the time of writing, the software can support smoking, alcohol consumption, exercise and weight/BMI entity lookups. The script has been designed in such a way that most R users can extend the code and provide unique behaviour for a lookup entity of interest. We will continue to support the scripts, adding in new entities and medcode descriptions as our research requires. This script is part of a family of R packages and scripts currently under development.
Zenodo: CPRD_Additional_Clinical, http://doi.org/10.5281/zenodo.39896346.
This project contains the following underlying data:
R script available from: https://github.com/acnash/CPRD_Additional_Clinical
Archived R script as at time of publication: http://doi.org/10.5281/zenodo.39896346.
License: MIT
RRID:SCR_018959
The script also holds medcode descriptions taken from the CPRD data dictionary for several entities. Samples of the clinical data and additional clinical data have not been provided as access to this data is only permitted following execution of the appropriate license agreements. If access to any of this data is required, we can provide medcode lists and the criteria used to extract the dataset (ISAC project 17_255R2) used to create this software, and we recommend contacting CPRD directly.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new method (or application) clearly explained?
Partly
Is the description of the method technically sound?
Yes
Are sufficient details provided to allow replication of the method development and its use by others?
No
If any results are presented, are all the source data underlying the results available to ensure full reproducibility?
No
Are the conclusions about the method and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Health Data, Machine Learning, Text Mining
Is the rationale for developing the new method (or application) clearly explained?
Partly
Is the description of the method technically sound?
Yes
Are sufficient details provided to allow replication of the method development and its use by others?
No
If any results are presented, are all the source data underlying the results available to ensure full reproducibility?
No
Are the conclusions about the method and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genomics, Bayesian, Machine learning.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 11 Sep 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)