Keywords
Image processing, image analysis, automation, OMERO, ImageJ, Fiji
This article is included in the NEUBIAS - the Bioimage Analysts Network gateway.
Image processing, image analysis, automation, OMERO, ImageJ, Fiji
Modern microscopy, through new systems like light-sheet or high-throughput microscopes, is generating a vast amount of complex data that needs to be analysed. These data can be large in size or in number. Furthermore, for the purpose of reproducible quantitative analysis, these data should be batch-processed by a standardized system, that can be easily shared and reused. Some batch systems already exist such as CellProfiler1, ICY2 protocols or ImageJ macros3. However, these systems may require some programming knowledge or time to set up for inexperienced users or are not yet fully multi-dimensional.
In the last 10–20 years, and more recently with deep-learning methods, a lot has been accomplished in the field of image processing, especially for image segmentation. However, there is no real standardization for image analysis protocols. Arganda-Carreras and Andrey4 designed a first version of a systematic image analysis pipeline. Furthermore, due to the recent advances in fast volumetric microscopy, more and more data are produced, but it lacks a systematic way of organizing raw data and subsequent analysed data and results. With the spread of database systems such as OMERO5, more and more imaging facilities and labs are storing their data in a more organized fashion.
We developed TAPAS (Towards an Automated Processing and Analysis System) as a system for describing and exchanging processing workflows. The protocols are simple text files comprising a linear sequence of commands. An extensive set of 60 commands is already available, mostly based on the 3D ImageJ Suite6,7. The design of the protocol allows simplified tracking of processed data and quantitative results, by using keywords to design the image data, such as ?image?.
TAPAS is focusing on data organization rather than complex segmentation or analysis algorithms. TAPAS focused originally on data stored on an OMERO database, by allowing to retrieve, perform classical segmentation procedures and analysis, and push back the results, both images and tables, to the database. Data on OMERO are, by design, organized by user, then projects and datasets. In TAPAS the current analysed image is simply referred by the keyword ?image?, and the corresponding project and dataset the data belongs to by ?project? and ?dataset? respectively. Subsequent processed data are then referred as ?image?-processing, for instance ?image?-nucleus for the result of nucleus segmentation. Similarly, additional datasets can be created such ?dataset?-labels to store the results of segmentation. The results tables can be stored using the name of the image as reference such as ?image?-nucleus-volume.csv. Results tables will be linked to the original raw image using OMERO attachments.
The system is implemented in Java, with a core library, including OMERO and BioFormats input/output utilities, and a plugins library including a comprehensive set of modules. Each module is generic as it will process a generic Image class, and each class will get an Image as input and will return an Image as output. Parameters are managed as simple String files, allowing flexible management of parameters, even for inexperienced Java programmers. The current system uses the ImageJ ImagePlus class as implementation for the Image class, however any other class can be used, allowing the use of any Java library.
The processing pipeline is constructed as an ordered list of processing classes, with their corresponding parameters. Since the classes are generic, a processor is also specified as how to process the image data, by default an ImagePlus processor is built. An experimental version of processor with a set of processing modules using the clearCLBuffer class has been tested and validated using the CLIJ system8.
TAPAS is java-based and works with the ImageJ/Fiji system, the use of a OMERO database is optional. TAPAS is designed to work with a database; either OMERO or a local database. A local database is a folder organized, not unlike OMERO, as projects, datasets, and finally images. We also add an attachments' folder to store the results tables. Having a completely similar organization between OMERO and a local database allows to have an exact same protocol to run using an OMERO database or a local database. A typical workflow of the system is presented in Figure 1.
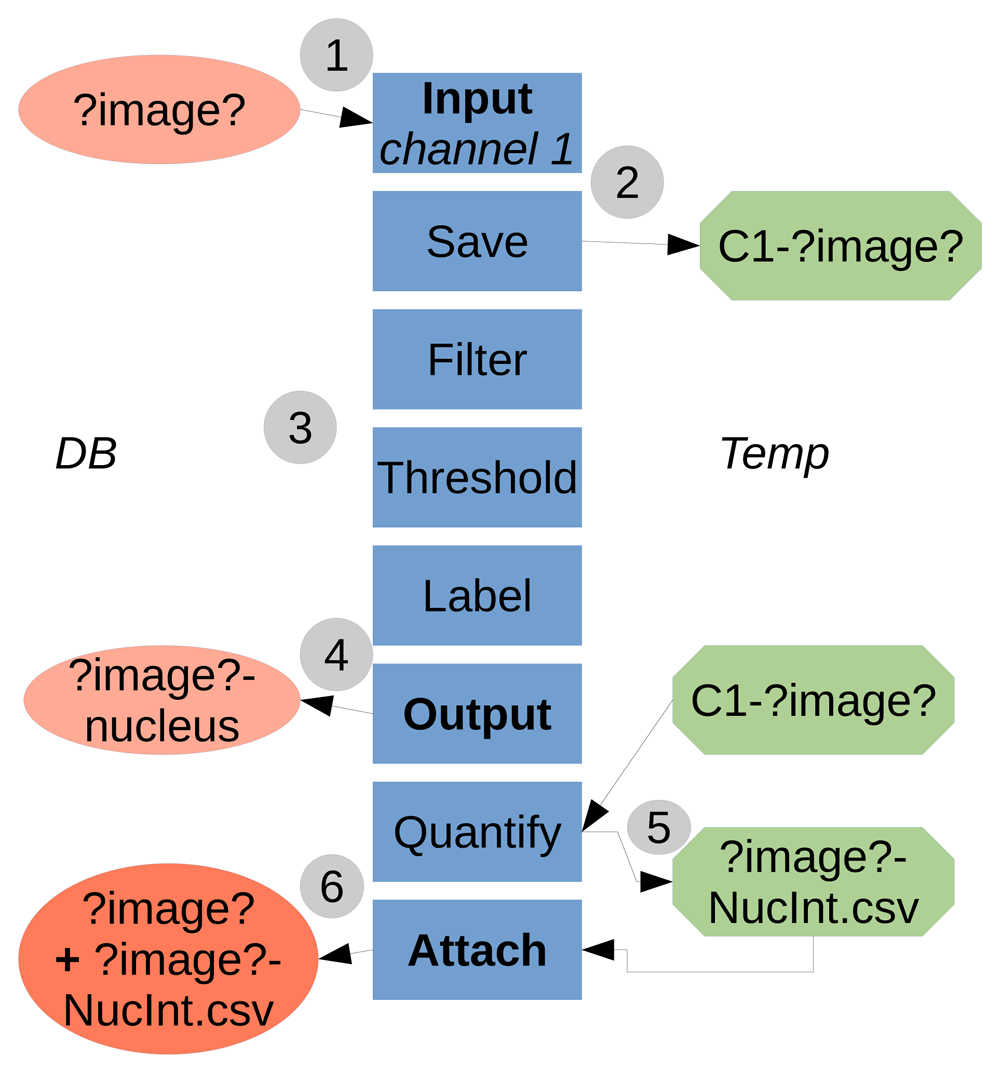
1) The data to be processed is input into the processing pipeline from the Database (either OMERO or local). ?image? is a keyword used to refer to the image name. The names in boxes refer to the module names. 2) The necessary data to be used later is saved locally, in a temporary folder (home folder, ImageJ/Fiji folder or system temporary folder). Here we saved the raw data for channel 1. 3) The data is processed, here a classical pipeline consisting of filtering, thresholding and labelling. 4) The resulting labelled data is output to the Database, here the labelled structure for channel 1 is the nucleus. 5) The previously saved raw data is used as parameter to quantify intensity inside the labelled nuclei. The results table is saved first in a file locally. 6) The results table file is then attached to the original processed image. The temporary saved data (raw data for channel 1 and results table file) can be then deleted within the pipeline or manually.
The system separates the data to be processed from the processing pipeline. Firstly, the list of image data to be processed is built, each image data to be processed is identified by its project, dataset and name (either on OMERO or on a local DB), and by the channel and frame to be processed. Second, the processing pipeline file is to be selected. After clicking run, the system will process the images sequentially, displaying information for each module, and the final processing time per image. Raw data will be pulled from the database and processed and analysed data will be pushed back to the database.
We propose a simple TAPAS menu that will display in an organized manner the list of available modules with their corresponding category and documentation. After selecting a module, the list of parameters will be displayed, the user can then manually enter the parameters values, and the corresponding processing pipeline text will be created.
TAPAS is a comprehensive system for data processing automation, relying on an extensive set of more than 60 modules for processing and analysis of multi-dimensional image data. An extensive documentation including the list of modules, various tutorials and links to the source code is available at https://imagej.net/TAPAS.
All data underlying the results are available as part of the article and no additional source data are required.
Software available from: https://imagej.net/TAPAS.
Source code available from: https://github.com/mcib3d/tapas-core/.
Archived source code at time of publication: http://doi.org/10.5281/zenodo.40911779.
License: GPL 3.0
This publication was supported by COST Action NEUBIAS (CA15124), funded by COST (European Cooperation in Science and Technology)
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: computational and system biology, image analysis, high-throughput screening and imaging, machine learning, in vitro cell models
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
No source data required
Are the conclusions drawn adequately supported by the results?
Yes
References
1. de Chaumont F, Dallongeville S, Chenouard N, Hervé N, et al.: Icy: an open bioimage informatics platform for extended reproducible research.Nat Methods. 2012; 9 (7): 690-6 PubMed Abstract | Publisher Full TextCompeting Interests: I do work on software solutions with similar purpose as TAPAS. I hope anyway that my review is appropriate and my feedback allows the authors to improve the manuscript.
Reviewer Expertise: Bio-Image Analysis, Computer Science
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 02 Jul 21 |
read | read |
|
Version 1 28 Oct 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)