Keywords
Achillea millefolium L., AMCSNPs, Drug delivery, Targeted treatment, Antibacterial, Antiurolithiatic
This article is included in the Nanoscience & Nanotechnology gateway.
Achillea millefolium L., AMCSNPs, Drug delivery, Targeted treatment, Antibacterial, Antiurolithiatic
Natural biopolymers are attractive products of living organisms as they serve a number of different applications for human health due to their biodegradability, such as vaccine delivery, drug development, and food preservatives1. Chitosan (CS) is a natural biopolymer and a derivative of chitin. It is obtained from different sources of chitin and differs on the basis of its degree of deacetylation2. In the last few years, CS nanoparticles (CSNPs) have drawn much attention due to their biodegradability, biocompatibility, quantum size effects, large surface to volume ratios, and their simple and inexpensive production3–5. Different biological activities of CSNPs have been reported, such as antimicrobial, antioxidant, anticancer6, drug delivery, tissue engineering, carbon nanotube, food preservative, and purification of water7. CSNPs successfully used in drug delivery for treatment of various diseases, including ocular drug delivery8, per-oral drug delivery9, nasal drug delivery10, pulmonary drug delivery11, mucosal drug delivery12, gene delivery13, buccal drug delivery14, vaccine delivery15, vaginal drug delivery16, and cancer therapy17 have been reported. Achillea millefolium is a perennial herbal aromatic plant belonging to the family Asteraceae with characteristically finely divided leaves and inflorescence in corymbose cluster. It has been reported to have different medicinal activities, including antibacterial and diuretic18,19. The current study was designed to evaluate the potential of CSNPs of A. millefolium (AMCSNPs) as an effective alternative of targeted drug delivery and treatment of various diseases, including bacterial infections specifically urolithiasis.
A. millefolium was collected from Pathanteer Village, Mendhar Tehsil, Poonch District(Jammu and Kashmir, India; GPS coordinates 33° 39’ 40” N- 74° 11’ 11” E) and identified by Raw Materials Herbarium and Museum (RHMD), National Institute of Science Communication and Information Resources (NISCAIR), Pusa with reference IDNISCAIR/RHMD/consult/2018/3293-94. 5g powder of inflorescence of A. millefolium was extracted in 50 ml of water by maceration at 90° C using water bath.
Crude extract of the plant was evaporated using rotary evaporator (Khera KI- 102), which resulted in the semi solid form of extract. This was then weighed and dissolved in a known amount of solvent for making a stock concentration of the plant extract. Different concentrations were made by serial dilution.
Strains of Bacillus subtilis (MTCC 441) and Pseudomonas aeruginosa (MTCC 1688) were procured from MTCC Chandigarh. All the chemicals used (chitosan, acetic acid, sodium tripolyphosphate (STPP), Tween-80, calcium chloride, sodium oxalate, tris buffer and NaCl) were of good quality and purchased from Fisher Scientific International, Inc.
CSNPs were prepared by ionic gelation method. 10ml 0.1% chitosan solution was made in 1% acetic acid with different percentages of the cross-linking agent STPP (0.5%, 1%, 1.5% and 2%). 5ml of STPP was added drop wise to the chitosan-acetic acid solution, which was magnetically stirred at room temperature. An opalescent color was observed, and stirring was continued for 60 min.
To obtain AMCSNPs, variable concentrations of plant extract (5%, 10%, 15% and 20%) were added to the 10 ml chitosan solution by magnetic stirring prior to adding the 5 ml STPP drop wise. This solution was stirred for a further 2 h followed by centrifugation at 10000g for 10 min and then the AMCSNPs were washed three times with distilled water. The pH of the nanoparticles was maintained at 4.8, and 12 drops of 1% Tween-80 was used to prevent agglomeration.
Percentage encapsulation efficiency of each concentration of extract was determined using the following formula20–22:
Encapsulation efficiency (%) = (total amount – free amount/total amount) *100.
UV-Vis spectroscopy using SPUV-1000 spectrophotometer attached to Mwave professional software 2.0 (or any software used to obtain the UV- Vis absorption spectra) and spectrum between 200 nm-700 nm was obtained for determining the main absorbing region. FTIR (Fourier Transform-Infrared) Spectroscopy using spectrometer (Brukers) in the range of 1000 cm-1–3500 cm-1 to identify the peaks of main functional groups, DLS (Dynamic Light Scattering) in the range between 0 nm to 1000 nm using zetasizer Nano ZS90 (Malvern Instruments Ltd., UK) at room temperature for particle size distribution and TEM (Transmission electron microscopy) at an accelerating voltage of 200 kV using Tecnai G2 30U-twin kV Ultra-twin microscope to study the morphology.
Primary culture of bacteria was obtained from lyophilized culture by inoculating in LB broth, which was incubated in an incubator shaker at 120 rpm and 37°C for 12–16 h. Pure colonies of each bacterium were obtained from primary culture by streak plate method using LB agar plates, which were inoculated in LB broth, incubated in an incubator shaker at 120 rpm 37°C for 12–16 h. Absorption of bacterial culture was adjusted to 0.1±0.02 at 600nm using SP-UV1000 spectrophotometer to reach the concentration of 108 CFU/ml for final use, which is equal to 0.5 McFarland standards, as previously performed in the literature23–25 to obtain a similar concentration of each bacterium. Each reading was taken thrice.
Antibacterial screening of AMCSNPs was done using well-diffusion method31. 1.5% LB agar plates were used and a 5mm cork-borer made four wells in each plate. 20 µl of B. subtilis and P. aeruginosa culture was added to the plates and spread using a glass spreader. 100 µl of AMCSNPs was poured in the wells. Plates were sealed with parafilm, incubated at 28° C for 12–16 h and the zone of inhibition (ZOI) recorded.
Nucleation and aggregation assays were used to determine the antiurolithiatic potential of AMCSNPs.
Nucleation assay: The method of Hennequin et al.27 was used with some minor modifications. Solutions of calcium chloride and sodium oxalate were prepared at a final concentration of 3mmol/l and 0.5mmol/l, respectively, in a buffer containing Tris 0.05mol/l and NaCl 0.15mol/l at pH 5.5. A total of 1.9 ml of calcium chloride solution mixed with 200 µl of AMCSNPs was incubated for 30 minutes in a 37° C water bath. Crystallization was started by adding 1.9 ml of sodium oxalate solution. Equal volume of water has used for a control instead of AMCSNPs. The optical density of the solution was recorded at 620 nm for 420 sec using spectrophotometer SPUV- 1000.
% Inhibition = {(Abs. Control- Abs. Sample)/ Abs. Control} * 100
Aggregation assay: The method of Hess et al.28 was used with some minor modifications. `Seed' CaOx monohydrate (COM) crystals were prepared by mixing calcium chloride and sodium oxalate at 50mmol/l. Both the solutions were equilibrated in a 60° C water bath for 1h and then cooled at 37° C overnight. The crystals were harvested by centrifugation at 10000g and then evaporated at 37° C. COM crystals were used at a final concentration of 0.8 mg/ml, buffered with Tris 0.05 mol/l and NaCl 0.15 mol/l at pH 5.7. A total of 1 ml of AMCSNPs were added in a test tube to 3 ml COM crystal solution and incubated at 37°C. Equal volume of water was used for a control instead of AMCSNPs. Absorption at 620 nm was recorded at different time intervals (30 min, 60 min, 90 min, and 120 min).
% Inhibition = {(Slope Control- Slope Sample)/ Slope Control} * 100
Statistical analysis was performed using Microsoft Excel 2007. One-way ANOVA was used followed by t-test to determine the significant difference of antibacterial activity between different samples and regression analysis was used to plot graphs of nucleation and aggregation assays.
On addition of STPP to chitosan solution, an opalescent color was observed, which indicates the formation of CSNPs. Different concentrations of STPP (0.5%, 1%, 1.5%, and 2%) were used for nanoparticle preparation and 1.0% STPP was found to be most suitable with sharpest peak shown by UV spectroscopy, indicating the most CSNPs made (Figure 1). Therefore, 1.0% STPP was used to obtain AMCSNPs. Different percentages of A. millefolium water extract (5%, 10%, 15% and 20%) in 0.1% of chitosan solution were used to make AMCSNPs and excellent loading efficiency was observed, i.e. 94%, 94.7%, 94.7%, and 95.2% for 5%, 10%, 15% and 20% A. millefolium respectively. A standard graph for absorbance of A. millefolium extract at 417 nm (maximum absorption verses concentration of extract) was obtained. The amount of loaded extract was determined using the standard graph as a decrease in the absorption values of the supernatant of AMCSNPs indicated the loading of extract of the nanoparticles. Loading efficiency was calculated using the above encapsulation efficiency formula for each concentration. Hence 20% AMCSNPs have been used for further analysis.
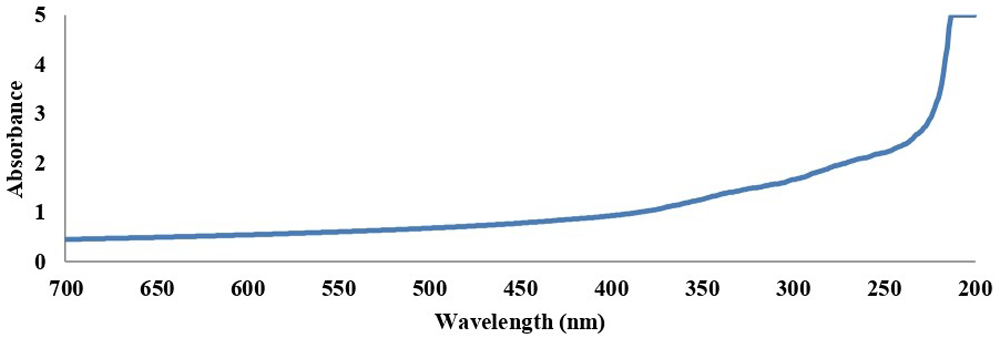
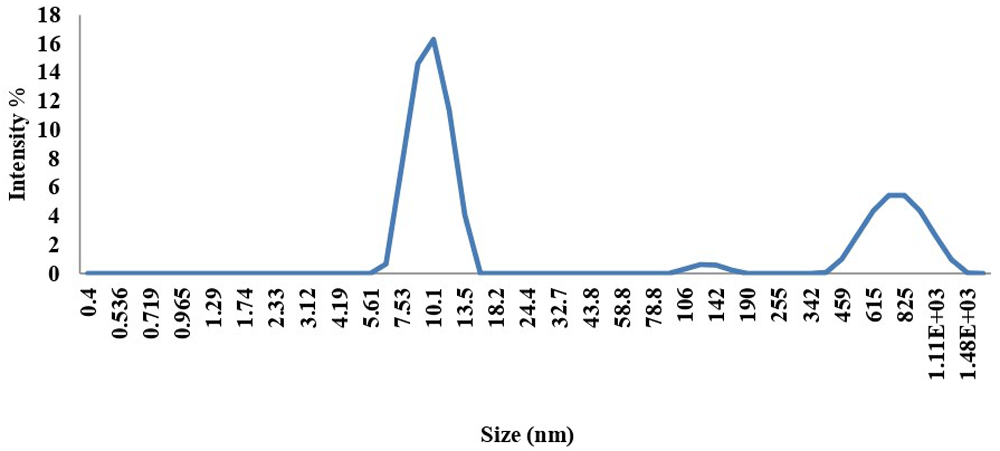
A broad absorption band between 200 to 300 nm was shown for the UV spectrum of AMCSNPs (Figure 2). FTIR spectra of CS showed peaks at 3324.15, 2153.28, 1638.72 and 1279.29; FTIR spectra of CSNPs showed peaks at 3317.48, 2139.29 and 1638.46; and FTIR spectrum of AMCSNPs showed peaks at 3281.73, 2163.36 and 1636.78 (Figure 3). DLS revealed the size range of nanoparticles with Z average of 118nm having characteristic peaks at 10 nm, 122 nm and 712 nm, and highest intensity was recorded at size 10nm (Figure 4). TEM was used to study the morphology of the nanoparticles, which revealed a spherical shape with smooth surface. TEM also revealed the size of AMCSNPs: <100 nm with smallest size of 4.15 nm (Figure 5).
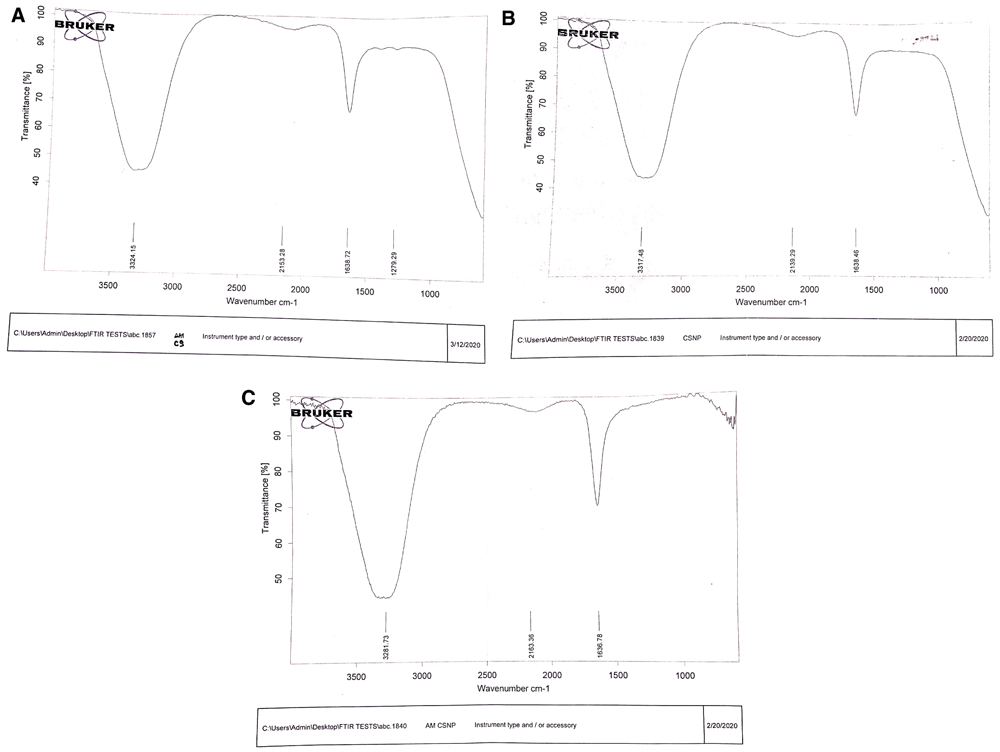
Fourier Transform-Infrared graph of chitosan (A), chitosan nanoparticles, (B) and Achillea millefolium chitosan nanoparticles (C).
AMCSNPs exhibited excellent antibacterial activity against both Gram-positive B. subtilis and Gram-negative P. aeruginosa. AMCSNPs showed three-times the increase in antibacterial activity as compared with A. millefolium extract only (control); ZOI increased from 10 mm to 30mm against both B. subtilis and P. aeruginosa with a statistically significant difference between A. millefolium, CSNPs and AMCSNPs (Figure 6).
AMCSNPs showed significant antiurolithiatic activity with 68% inhibition in the aggregation assay and 51.26% inhibition in the nucleation assay as compared to 55.132% and 9.09% inhibition by A. millefolium extract (control). In the nucleation assay, the % inhibition is nearly equal in the case of A. millefolium and AMCSNPs, but CSNPs did not show any inhibition. In the aggregation assay there is a significant increase in % inhibition with 9.09%, 63.63% and 68% inhibition by A. millefolium, CSNPs and AMCSPs, respectively, (Figure 7).
For characterization of nanoparticles, different techniques used in the literature including UV- Vis spectroscopy, FTIR spectroscopy, DLS and TEM29,30. In our study, a UV-Vis absorption band for AMCSNPs of 200-300 nm indicates the presence of a CO group in the CSNPs, as reported by Vaezifer et al.31. A shift of FTIR peaks from 3317.48, 2139.29 and 1638.46 for CSNPs to 3281.73, 2163.36 and 1636.78 for AMCSNPs indicates that the loading of A. millefolium into the CSNPs, as reported by Khan et al.32. Our DLS results are comparable to the average size of CSNPs reported in literature, i.e. 189 and 197 nm by Khan et al.32, 216 nm by Agarwal et al.33, and size range of 135–729 nm by Rasaee et al.34 and 6.5-1331.2 nm by Iswanti et al.35. TEM of AMCSNPs in our study revealed a spherical shape with a smooth surface, which was also reported by Da Silva et al.21.
Chitosan is a positively charged macromolecule, which interacts with the negatively charged microbial membrane, and results in the breakage of intracellular components. Chitosan acts as a chelating agent and limits toxin production and microbial growth36–38. Antibacterial screening of Ocimum basilicum CSNPs against E. coli and Bacillus vallismortis have been reported by Rasaee et al.34, chitosan-tripolyphosphate nanoparticles against Staphylococcus aureus and P. aeruginosa have been reported by Bangum et al.39, and against phytopathogens of tomato Xanthomonas and Erwinia strains by Oh et al.29. Gallic acid-chitosan conjugates have been reported to inhibit the formation of calcium oxalate crystals by Queiroz et al.40. Antiurolithiatic activity of Aerva lanata chitosan nanoparticles at 0.8 µg/ml concentration through nucleation assay have been reported by Chandirika et al.41 and Tridax procumbens by Chandirika et al.42. In our study, AMCSNPs showed excellent antibacterial activity against both B. subtilis and P. aeruginosa, and significant antiurolithiatic activity at aggregation stage of urolithiasis.
Figshare: Chitosan nanoparticles of Achillea millefolium L., https://doi.org/10.6084/m9.figshare.12936293.v343.
This project contains the following underlying data:
Output files of chitosan nanoparticles with different concentrations of STPP
Raw data for Figures 1, 2 and 4
Unedited and uncropped FT-IR graphs and TEM images of AMCSNPs
ZOI of antibacterial activity of AMCSNPs
Absorption values of antiurolithiatic activity AMCSNPs
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
The authors are thankful to the Principal, Dr. Manoj K. Khanna, Ramjas College, Prof. S.B. Babbar, Head, Prof. K.S. Rao, and Prof. Veena Agrawal, Department of Botany, University of Delhi, Delhi for providing necessary facilities and encouragement during the course of investigation.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Stress Biology, Secondary Metabolite Biotechnology, Proteomics
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Plant Biotechnology, Genomics, Molecular Genetics, Bioinformatics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 04 Nov 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)