Keywords
Alzheimer’s Disease, Down Syndrome, Behavior, Memory, Learning
Alzheimer’s Disease, Down Syndrome, Behavior, Memory, Learning
Amyloids are peptide or protein aggregates that form from the misfolding of normally soluble proteins, which then stick together due to their chemical properties and accumulate in extracellular compartments and organs1. Amyloids form fibrous structures and plaques that are highly insoluble, resistant to degradation, and are involved in several diseases such as Alzheimer’s disease (AD), Down syndrome (DS), spongiform encephalopathies, and type II diabetes2,3. The amyloid plaques associated with AD are formed from peptides derived from the mis-processing of APP, a protein that typically resides around nerve cells2. The toxic peptide fragments are called beta amyloids. In AD, the amyloid plaques deposit in brain tissue, destroy neuronal connectivity, disrupt signaling at synapses, and eventually result in nerve cell death, tissue loss, and a reduction in brain mass2. Smaller aggregates of beta-amyloid and not the plaques themselves trigger the immune system and inflammatory processes4.
Early onset AD that runs in families is linked to the APP and PSEN1/PSEN2 genes5. A mutation in one of these three genes may cause AD to develop early whereas the more general form of the disease, late onset, is typically linked to the APOE gene6. PSEN1/PSEN2 are transmembrane proteins that are the catalytic subunit of gamma secretase, the enzyme responsible for cleaving APP. Mutations in the PSEN genes may result in the abnormal cleaving and processing of APP to smaller toxic beta amyloid fragments which aggregate and accumulate7,8.
People with Down Syndrome (DS) are born with an extra copy of chromosome 21 (Chr 21) and many of these individuals develop AD as they age9. This is due at least in part to the extra copy of the APP gene located on Chr 21. By the age of 40, most people with DS have amyloid plaques which disrupt brain cell function and increase their risk for AD3. About half of the people with DS develop AD and the associated dementia around 50 to 60 years of age, which is about the age at which the hereditary form of AD, early onset AD, manifests10. Duplication of APP alone, in the absence of Chr 21 trisomy, is another cause of early onset AD11,12 making it likely that having three copies of APP is important in the development of AD in DS.
In both early and late onset AD the clinical symptoms include dementia, memory decline, and the inability to retain recent information or store new memories13. As the disease progresses, AD individuals may exhibit problems with language, reasoning, decision making, executive function, mood swings, aggressive behavior, and apathy. Late stage symptoms of AD may result in seizures, hypertonia myoclonus, incontinence, and mutism14. Death commonly occurs from general inanition, malnutrition, and pneumonia.
Memory loss and forgetfulness, which is typical in AD individuals, is less pronounced in people with both DS and AD. This may in part be a floor-effect due to the memory deficit already present in DS individuals15–17. Studies report an impairment in verbal short-term memory (example: serial order of a list of words) relative to visuo-spatial memory (manual selection in serial order of locations) and deficits in explicit long-term memory18. Also in individuals with DS there is evidence of hippocampal dysfunction and deficits in prefrontal systems as compared with mental age-matched controls19. In DS individuals, the hippocampal volume is reduced prior to the onset of dementia, and these reductions were found to relate to memory mainly due to the loss of neurons and neuronal volume as a result of neurofibrillary tangle formation20.
In this study we investigate the relationship between AD and DS through integrative geneset analysis of genes derived from peptides associated with amyloid plaques found in AD and DS individuals, Chr 21 genes, AD risk factor genes, and differentially expressed genes (DEX) identified through a transcriptome analysis of DS individuals for both the dorsal frontal cortex (DFC) and cerebellar cortex (CBC).
All genesets used in this study are presented in Extended data Workbook 121. The Chr 21 geneset was obtained from NCBI Gene. A total of 250 unique gene IDs were obtained at the time of manuscript preparation (September 1st, 2020). The AD-DS geneset, consisting of 292 genes, was obtained from GeneWeaver using “Alzheimer’s Down Syndrome” as the search term. The geneset was originally generated via gene2mesh v.1.1.1 (updated: 2019-01-07) from Medical Subject Headings (MESH Terms) GS236695 • [MeSH] Amyloid beta-Peptides: D016229. The AD risk factor geneset is comprised of 279 genes, many of which were identified and/or confirmed through a large scale GWAS of 71,880 clinically diagnosed AD and AD-by-proxy cases and 383,378 controls22.
The DEX genesets for the DFC (842) and CBC (570) were obtained from The Down Syndrome Developmental Brain Transcriptome database. Human Brain Transcriptome, Department of Neurobiology Yale University School of Medicine which is a publicly accessible database containing searchable differential gene expression information of transcriptome data in developing and adult DS versus control human brains. The data was generated from 15 sets of a DS and a matched control brain each. The specimens ranged from embryonic development to adulthood23.
Common genes among the AD-DS, Chr 21, DEX DFC, DEX CBC and AD risk factor genesets were assessed using venn diagram analysis (http://www.interactivenn.net/) and visualized with the UpSet Library in RStudio, R Version 4.0.2.
Keyword categories were used to evaluate the genesets. The keyword categories were chosen based on the major phenotypes associated with AD and DS. The terms used were: aging, Alzheimer's disease, amyloid; apoptosis, behavior cholesterol, circadian, cognition; Down Syndrome, face, fibril, immune, inflammation, insulin, learning, leptin, memory, muscle, myelin; obesity, sleep, speech, and tau.
Gene ontology characterization of the genesets was performed in both DAVID and the Gene Ontology database for Biological Process (BP). The Benjamini corrected P-value was used to determine enrichment significance. Functional information based on GO annotations for the genes associated with a keyword search term related to AD and DS were identified and noted.
Gene Ontology pathway enrichment was used to further characterize the AD-DS and Chr21 genesets in order to obtain a broader overview of collective gene function. The Benjamini-corrected P-value was used to determine significance.
The APP protein-protein interaction network was built in STRING (version 11.0), based on experimentally validated interactions. The combined scores for the interactions are computed by combining the probabilities from the different evidence channels and corrected for the probability of randomly observing an interaction. First and 2nd shell interactions are included in the network. The network was exported from STRING and analyzed in Cytoscape (version 3.7). Network bottlenecks and clusters were identified with Cytoscape plugins CytoHubba (version 0.1) and MCODE (version 1.6.1), respectively.
The number of common genes among all of the 5 genesets (AD-DS, Chr 21, AD risk factors, DEX DFC, and DEX CBC) along with the gene names and Gene Ontology classifiers are shown in Figure 1 and Extended data Workbook 224. The AD-DS, Chr 21 and AD risk factor genesets overlap by eight genes: APP, BACE2, COL18A1, DYRK1A, RCAN1, SOD1, SYNJ1, and S100B (Figure 1). BACE2 encodes an integral membrane glycoprotein that cleaves the APP protein into amyloid-β, a critical step in the cause of AD and DS. COL18A1 encodes the alpha chain of type XVIII collagen. It is associated with vascular deposits and senile plaques in AD brains25. The DYRK1A gene product can phosphorylate APP and alter the protein’s stability and the formation of amyloid-β26,27. Increased RCAN1 expression is associated with neuronal death and Tau hyperphosphorylation, as well as neurofibrillary tangle formation in DS and AD individuals28.
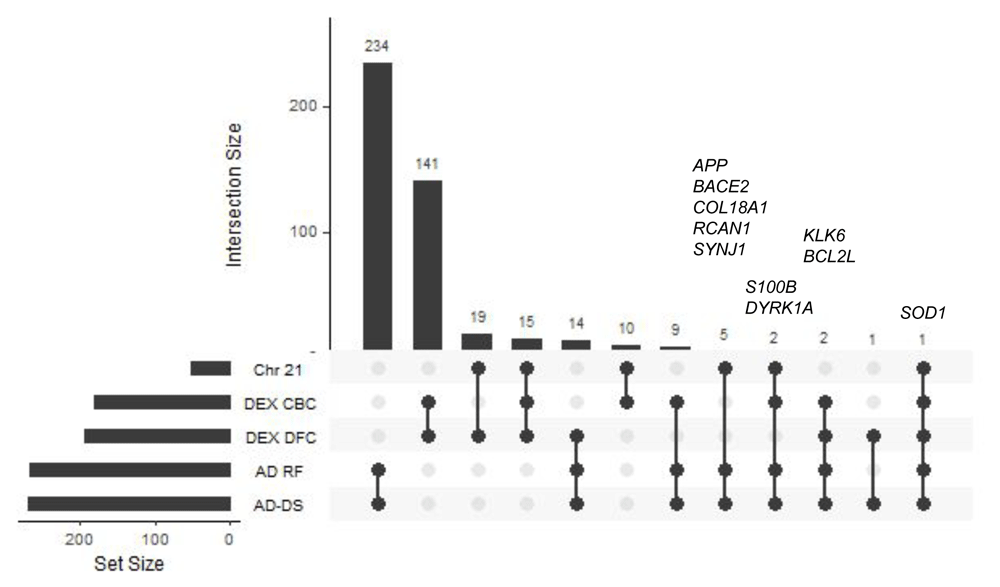
UpSet plot showing geneset overlap highlighting gene content similarity between the AD-DS, Chr21, AD risk factors DEX DFC, and DEX CBC genes.
SOD1 is the only gene present in all of the genesets. SOD1 is associated with apoptosis and oxidative stress29. The extra copy of SOD on Chr 21 results in increased gene expression and increased production of H2O2 which is believed to underlie many of the DS-related pathologies29. SOD1 is also associated with neurodegeneration in amyotrophic lateral sclerosis and AD30,31. SYNJ1 encodes a lipid phosphatase that is involved in autophagosomal/endosomal trafficking and synaptic vesicle recycling. Its dysfunction has been linked to several neurodegenerative diseases, including AD and DS32. S100β belongs to a family of cytokines that are strongly associated with activity underlying AD related pathologies such as APP processing, protein inclusion formation, and Tau post-translational modifications. S100β is also linked to DS. S100β levels are increased in neuronal progenitor cells of DS patients33 and in human induced pluripotent stem cells derived from DS patients34. Two additional genes, KLK6 and BCL2L, are shared among the AD-DS, AD risk factors, DEX DFC and DEX CBC genesets. KLK6 has been proposed as a biomarker for AD35. BCL2L is located on the outer mitochondrial membrane and is a negative regulator of apoptosis36.
Each of the genesets were evaluated for association with AD and DS related phenotypes (Figure 2 and Extended data, Workbook 337). The keyword categories shared among all genesets are muscle, immune, insulin, glucose, behavior, oxidation and heart. The AD-DS geneset has a high frequency of genes associated with most of the keyword categories. The largest represented categories are: AD, muscle, inflammation/immune system, insulin, amyloid, behavior, aging, learning/memory, circadian processes and face/facial features. There were no genes directly associated with DS. For the Chr 21 geneset, unlike the AD-DS geneset, there were very few genes associated with the keyword categories. The highest frequency categories are immune, muscle, aging, behavior and insulin. Three genes are connected to AD (NDUFV3, APP and BACE2) and one with DS (DSCAM).
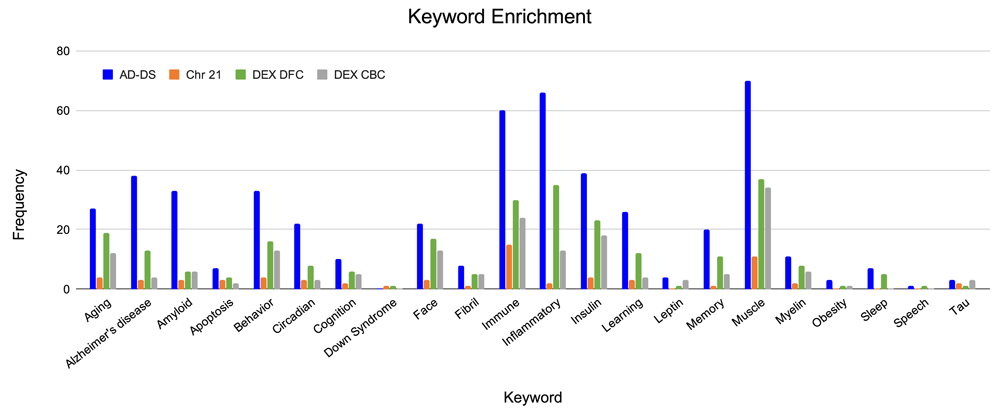
Identification of genes associated with terms relating to AD and DS based on gene ontology term classification. AD-DS genes: blue, Chr 21 genes: orange, DEX DFC genes: green, DEX CBC genes: gray, X-axis, keyword categories; Y-axis, frequency of occurrence in the geneset.
The enriched keyword categories for the DEX DFC are very similar to the results obtained for the AD-DS geneset: muscle, inflammation/immune system, insulin, aging, face/facial features, behavior, AD, and learning/memory. There are 13 genes directly associated with AD (NDUFS2, APAF1, BACE1, CACNA1F, COX5B, COX6A2, GRIN1, GRIN2A, LPL, PLD3, PSEN1, RYR3, UQCRC1) and one gene associated with DS (DSCR9). For the DEX CBC geneset the most representative categories are again similar to the AD-DS geneset as well as the DEX DFC geneset: muscle, immune/inflammation, insulin, behavior, face/facial features, aging and amyloid. There are four genes directly linked to AD (ATP5H, APOE, APAF1, RYR3). There are no genes directly associated with DS.
Given that behavioral phenotypes are highly shared between AD and DS, the specific types of behaviors identified from the keyword enrichment were evaluated more in depth. The AD-DS geneset has a large number of behavior related genes and genes related to learning and memory: (Behavior 33, Learning 26, Memory 21). This observation is based on the GO results obtained for three random genesets of the same size: Behavior 7,2,1; Learning 0,1,1; Memory: 0,0,1. The behavior gene categories are diverse and include fear, locomotion, eating and feeding, addiction related (nicotine, cocaine, ethanol), social, and others such as circadian, mating, and response to pain. The learning categories include visual learning, associative learning, and also olfactory, motor, and nonassociative learning. The memory related categories are short-term and long-term memory, and in one instance, susceptibility to memory impairment (Figure 3).
Many of the significant BP enrichment classifiers for the AD-DS geneset are associated with cell death (P=3.01E-83,) apoptosis (P=1.30E-70) and inflammation/immune system (P= 1.65E-36). For the Chr21 geneset, the significant BP enriched terms are linked to keratin (keratinization, P=1.04E-37), skin (skin development (P=2.83E-29) and epithelium processes (P=3.19E-15) as well as tissue (P= 3.56E-14), organ (P=3.40E-09) and anatomical structure development (P=8.66E-09). The significant pathways associated with the AD-DS geneset are related to neurodegenerative disorders (AD P=3.1E-23, Parkinson’s disease (P=1.39E-04) and Huntington’s disease P=1.36E-07) as well as many signaling pathways linked to insulin (P=1.86E-09) and inflammation (Jak/Stat P=9.49E-04), Toll receptor (P=4.04E-10), Interferon-gamma signaling (P=8.90E-06). There were no significant pathways associated with Chr 21. All pathways are listed in Extended data Workbook 438.
The transcription profiles of the AD-DS and Chr 21 genesets were evaluated here and compared with the DEX genesets which were previously evaluated by Olmos-Serrano et al.23 (Extended data, Workbook 539). There are 64 transcription factors present in the AD-DS geneset. Several of these are directly associated with AD (GSK3B, IL1B, MAPK3/8/10/14, WNT1, WNT3A, KAT5 NOTCH1 and TNF), Tau (GSK3B and CLU) and amyloid (CD36, NLRP3, CLU, FOXO3, PARP1, PRNP). Of these, many are related to mitochondria processes (AKT1, CLU, GSK3B, HIF1A, MAPK3,8,10,14, MTOR, NFKB1, PPARGC1A, PARP1, PRNP, PRKCA, SIRT1, STAT3, SREBF2, UBB) and also inflammation, oxidative stress, and aging (TP53, STAT1/3, NFKB1, HIF1A, and NEF2L2).
For the Chr 21 geneset, 18 transcription factors were identified. RUNX1 which is associated with ossification40 and nervous system development41 observed comparable expression in a study comparing AD and DS brains. Gene variants of RUNX1 are associated with both AD and DS42. The OLIG1/OLIG2 transcription factors regulate oligodendroglial differentiation and myelination and neuron fate commitment43. In DS, due to the gene triplication, OLIG1/OLIG2 causes alterations in brain development44. OLIG2 is associated with the psychotic symptoms of AD and also schizophrenia45. Of the Chr 21 transcription factors, only one is associated with mitochondria—GABPA— which is involved in the activation of cytochrome oxidase expression and nuclear control of mitochondrial function46.
There is one common transcription factor between the AD-DS and DEX-DFC genesets: NFE2L2 (also known as NRF2), which is associated with the oxidative stress response with aging, spatial learning, memory, and neuro-inflammmation via regulation of antioxidant response elements47,48. NFE2L2/NRF2 regulates BACE1, the rate-limiting enzyme for amyloid-β peptide (Aβ) generation. NRF2 activation decreases production of BACE1 and BACE1 antisense RNA (BACE1-AS) transcripts and Aβ production and ameliorates cognitive deficits in animal models of AD49. Depletion of NFE2L2/NRF2 increases BACE1 and BACE1-AS expression and Aβ production and worsens cognitive deficits50.
There are two transcription factors common between the AD-DS and DEX-CBC genesets. MTOR has been identified as a key target for therapeutic intervention in AD because of its regulation of several key signaling pathways: phosphoinositide 3-kinase (PI3-K)/protein kinase B (Akt), glycogen synthase kinase 3 [GSK-3], AMP-activated protein kinase (AMPK), and insulin/insulin-like growth factor 1 (IGF-1)51. Both upstream and downstream components of mTOR signaling are associated with AD progression and pathogenesis. MTOR inhibits autophagic processes and contributes to amyloid β-peptide generation and/or clearance52. MTOR activation also contributes to aberrant hyperphosphorylated tau53. The other common TF is NFKB1 which is a key regulator of innate immunity and strongly associated with the inflammatory response involving cytokines and chemokines54. NFKB1 is also linked to aging and AD55,56.
An APP protein-protein interaction network was created to identify genes from the genesets evaluated in this study that are connected to APP through 1st and 2nd shell interactions. A total of 362 proteins make up the network (Extended data Workbook 657).
The APP protein interaction network overlaps by 48 genes with the AD-DS geneset, 41 with the AD risk factor geneset, 21 with the DEX DFC, 12 with the DEX CBC geneset and four with the Chr 21 geneset. The shared genes are highlighted in the network to visualize and forecast additional genes that are potentially involved in APP signaling and that are relevant to both AD and DS (Figure 4A). The top proteins that bridge (bottlenecks) the different sections of the network and that may signify information flow are: APP, ENSG00000259680 (a novel protein coding gene with similarity to immunoglobulin heavy chain variable region.), SHC1, DLG4, STUB1, KLC1, GFA1, CENPJ, and GNO1.
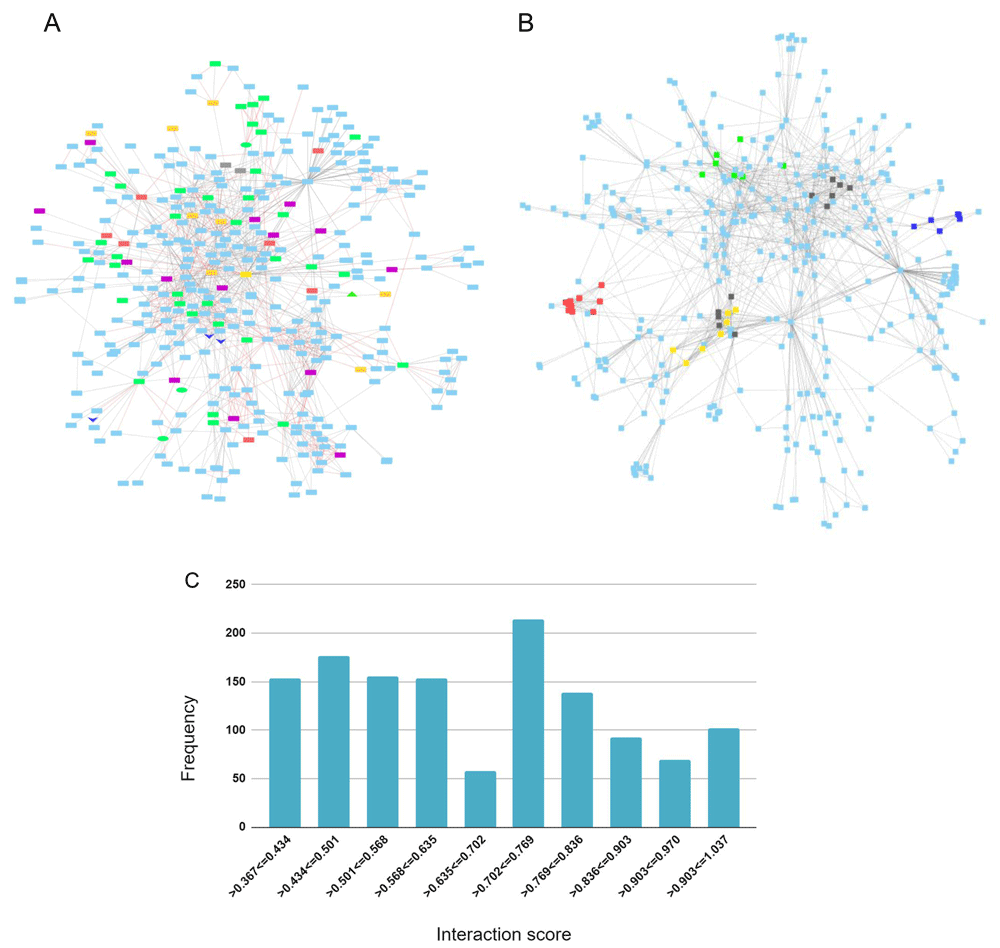
(A) Geneset overlap between 1st and 2nd shell interactions and the AD-DS, Chr 21, DEX DFC, and DEX CBC genesets. AD-DS genes unique: red; Chr 21 genes unique: gray; DEX DFC genes unique: purple; CBC genes unique: orange; AD risk factors (RF) and AD-DS genes shared: green; DEX DFC genes shared with RF & AD-DS genes: green oval; CBC and DFC shared genes: dark blue V; CBC genes shared with RF: green triangle; APP: yellow rectangle. (B) Interaction network gene clusters. Cluster 1: red – COP subunits, signalosome complex, development; Ubiquitin, Cluster 2: yellow – Tubulin, microtubules, motors, intracellular transport; Cluster 3: green – apoptosis, insulin signaling, ubiquitin, VEGFR growth factor signaling; Cluster 4: blue – Ubiqitin, autophagy; and Cluster 5: black – APP processing (PSEN, gamma secretase complex). (C) Distribution frequency for interaction score.
The APP network contains six major clusters (Figure 4B). Cluster 1: COP subunits, signalosome complex, development, ubiquitin; Cluster 2: TUBULIN, microtubules, motors, intracellular transport; Cluster 3: apoptosis, insulin signaling, ubiquitin, VEGFR growth factor signaling; Cluster 4: UBIQUITIN, autophagy; Cluster 5: APP processing (PSEN, gamma secretase complex); and Cluster 6: TUBULIN, microtubules.
The AD risk factor genes, Chr 21, and AD-DS genes are mostly dispersed throughout the network but a couple of areas in the network contain several connected AD risk factor genes. Predicted genes of interest based on their connectivity to these areas are METTL2B (tRNA methylation), IK (immune response), SAP18 (RNA splicing), QTRT1 (tRNA modification), APLP1 (Prion pathway), PRSS1( proteolysis, extracellular matrix digestion), ACSM1 (lipid metabolism), APBA2 (binds beta amyloid, synaptic transmission, and nervous system development). The validity of all of the interaction scores range from 0.4–1.00 and, for the most part, are uniformly distributed with 695 of the interactions falling in the low to mid-range of 0.4 and 0.7 and 617 falling in the mid to high-range of 0.7 and 1.0 (Figure 4C).
Genesets associated with AD, DS, and Chr 21 were evaluated to identify genes, transcription factors, and pathways that may shed light on the relationship between AD and DS. Genes common to multiple genesets are either directly involved in APP processing or in TAU post translational modification. Many of the genes associated with the amyloid plaques in AD and DS function in learning and memory. A network analysis of APP protein-protein interactions was used to analyze the topology and connectivity of the genesets and, based on interactions with common AD-DS genes and AD risk factor genes, provide the foundation to predict potential genes of interest. Genes that connect the network and represent information flow as well as regions of high interconnectivity are also of interest for follow up studies. Given the central role of APP related processes in the pathology of AD and DS, all of the proteins in the APP interaction network are either potential risk factors for AD or may contribute to disease progression in both AD and DS. Taken together, our findings indicate that oxidative stress, cell death/apoptosis, and inflammation/immune system processes likely underlie the pathogenesis of both AD and DS.
All data underlying the results are available as part of the article and no additional source data are required.
Figshare: Extended Data Workbook 1. Genesets: AD-DS, Chr 21, AD risk factors, DEX DFC and CBC, https://doi.org/10.6084/m9.figshare.13106693.v121.
Figshare: Extended Data Workbook 2. Common Genes: Gene overlap between the AD-DS, Chr 21, AD risk factors, DEX DFC and CBC genesets, https://doi.org/10.6084/m9.figshare.13106741.v124.
Figshare: Extended Data Workbook 3. Keyword Gene Enrichment: Enrichment of the AD-DS, Chr 21, AD risk factors, DEX DFC and CBC genesets, https://doi.org/10.6084/m9.figshare.13106750.v137.
Figshare: Extended Data Workbook 4. GO Terms and Pathways: Gene Ontology Biological Process terms and pathways associated with the AD-DS and Chr 21 genesets, https://doi.org/10.6084/m9.figshare.13106762.v138.
Figshare: Extended Data Workbook 5. Transcription Factors: TFs present in the AD-DS, Chr 21 genesets, https://doi.org/10.6084/m9.figshare.13106774.v139.
Figshare: Extended Data Workbook 6. APP Network File: APP protein-protein interaction network, https://doi.org/10.6084/m9.figshare.13106777.v157.
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Human genetics, Genetic epidemiology, Alzheimer’s disease, Down syndrome, Autosomal dominant forms of Alzheimer’s disease.
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Down syndrome and Alzheimer's disease.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Studies on the pathogenesis of AD in DS.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
|
Version 2 (revision) 25 Oct 21 |
||||
|
Version 1 05 Nov 20 |
read | read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)