Keywords
WGS, Genomics, mitDNA, mitogenome
This article is included in the Genomics and Genetics gateway.
WGS, Genomics, mitDNA, mitogenome
Brycon Müller & Troschel 1844 has 44 valid species and belongs to the order Characiformes. Species vary between medium and large size and are found in the hydrographic basins of South America and the Caribbean (Lima, 2017). Brycon, Chilobrycon, Henochilus and Salminus belong to the family Bryconidae (Abe et al., 2014). They occur in rivers of clean, flowing water, with a rock bed and high levels of oxygen (Botero-Botero & Ramírez-Castro,, 2011). Dependent on the riparian forest, for the most part they feed on allochthonous material, such as fruits, seeds and insects (Albrecht et al., 2009). Seven species of the genus are threatened, including Brycon nattereri, and the main reasons are fishing and environmental changes (Pavanelli et al., 2018). B. nattereri occurs in the Tocantins, Paraná and São Francisco river basins. Chromosome, molecular and population studies with specimens from tributaries of the last two river basins showed significant differences between individuals from different basins (Resende et al., 2020). B. nattereri would be the third complete mitochondrial genome sequenced in the genus, after B. orbignyanus and B. henni.
The two B. nattereri specimens used in this work was collected in the Salto Stream (19°27’54.9” S; 46°13’57.2” W) and Espinha Stream (19°02’32.8”S; 46°01’12.9”W) tributaries belonging to the Paraná and São Francisco River basins, respectively, from Minas Gerais State, Brazil.
After sampling, we brought the living specimens to the laboratory, euthanized them according to the technical standards of CONCEA (National Council for Control of Animal Experimentation of Brazil and CEUA/UFV) and the Animal Use Ethics Committee/Federal University of Viçosa (883/2019). We performed the sampling with licenses provided by SISBIO (Biodiversity Authorization and Information System; license number: 1938128) and SISGEN (National System for the Management of Genetic Heritage and Associated Traditional Knowledge; license number: A9FE946). We identified and deposited the specimens in the ichthyological collection of the Laboratory of Ecological and Evolutionary Genetics of the Federal University of Viçosa, Rio Paranaíba campus (voucher number: LaGEEvo-3928 and LaGEEvo-3847).
Total genomic DNA was extracted from liver and heart samples, as per the instructions of Invitrogen’s DNA extraction and purification kit (catalog number: K182002). After quality checking using Nanodrop (Lite Spectrophotometer ND-LITE-PR), Whole Genome Sequencing was performed by Illumina sequencing (San Diego, CA) at Novogene company, UK. The 2x150 raw reads were uploaded to Galaxy (Afgan et al., 2018), and trimmed to remove the adaptors and low-quality sequences using Trimmomatic Galaxy Version 0.38.0 (Bolger et al., 2014).
We assembled the mitogenome from raw reads on NOVOPlasty v3.7 (Dierckxsens et al., 2017) in a parallel cluster computer (64 Gb RAM) using the mitogenome of Brycon henni (GenBank KP027535.1) as seed. The mitogenome of Brycon from the Parana river basin was circularized on NOVOPlasty, but not from the São Francisco basin because of repeats on the D-loop region. Therefore, we uploaded the aligned reads to Galaxy and assembled de novo using SPAdes v3.13.0 (Bankevich et al., 2012). We blasted the sequences using megaBLAST implemented at NCBI obtained to be sure we had mitochondrial sequences (Altschul et al., 1990). We annotated the mitochondrial sequences on MitoAnnotator (Iwasaki et al., 2013) at MitoFish v. 3.60. We mapped as mitochondrial sequences in relation to the raw reads (after quality filtering) with Bowtie2 v. 2.4.1 (Langmead & Salzberg, 2012).
To validate our data, we aligned the CDS sequences of the two individuals of B. nattereri, B. henni (GenBank NC_026873.1), B. orbignyanus (GenBank NC_024938.1), Salminus brasiliensis (GenBank NC_024941.1), and Astyanax paranae (GenBank NC_031380.1) (outgroup) using MUSCLE (Edgar, 2004), followed by maximum likelihood analysis using MEGA X software (Kumar et al., 2018), with GTR model + gamma + invariant sites and 1000 bootstrap.
The mitochondrial genome of B. nattereri from Paraná and São Francisco river basin has the expected size for vertebrates, 16,837 bp (GenBank MT428073) and 16,752 bp (GenBank MT428074 access), respectively. Both specimens have a total of 37 functional genes (Figure 1), 13 DNA coding regions (CDS), two ribosomal RNAs and 22 transporter RNAs.
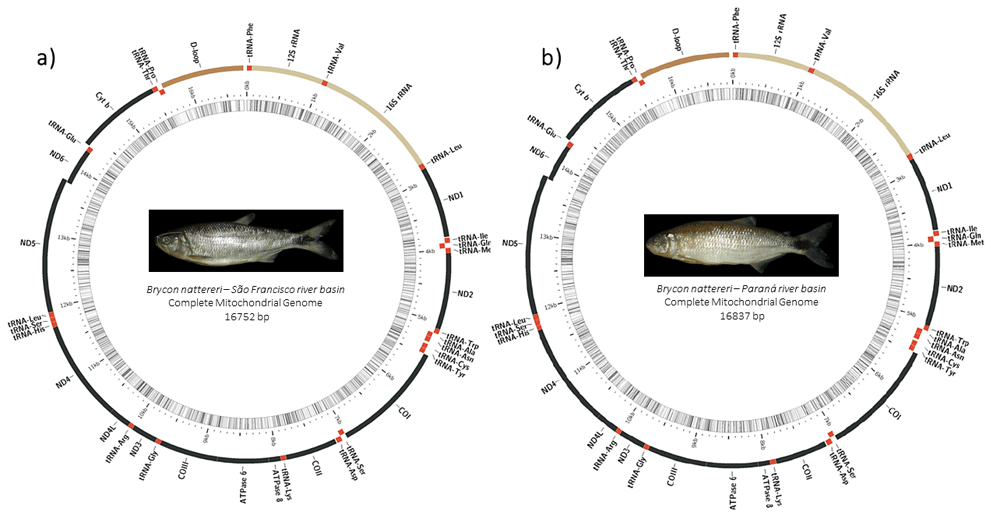
Illustrative diagram of the complete mitochondrial genome of Brycon nattereri from São Francisco (a) and Paraná (b) river basins, including genes encoding proteins, ribosomes and transporters, as well as their locations on the H and L strands.
The maximum likelihood dendrogram generated with our data and other complete GenBank mitochondrial genomes (Figure 2) rescued phylogenetic relationships according to literature (Abe et al., 2014; Hilsdorf et al., 2008).
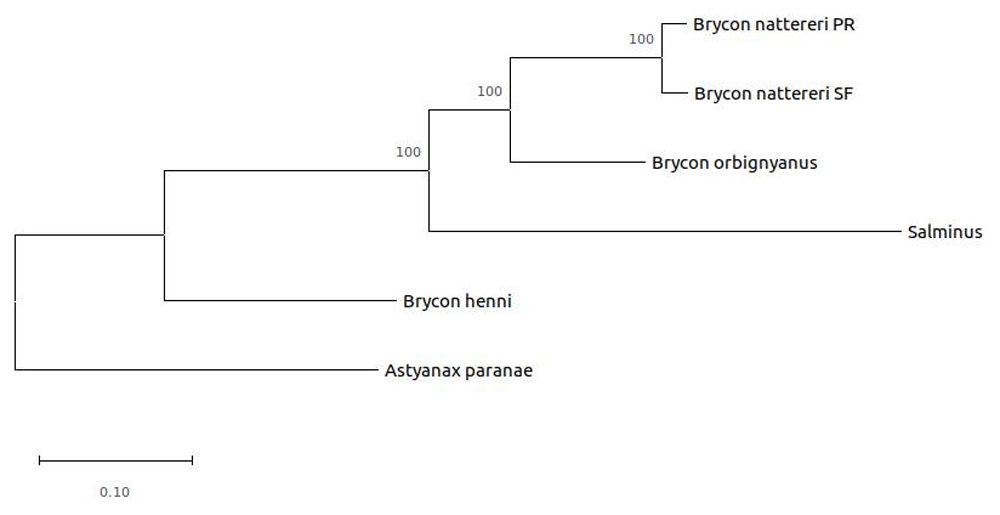
The evolutionary history was inferred by using the Maximum Likelihood method and General Time Reversible model. The tree with the highest log likelihood (-45932.69) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood approach, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 3.3372)). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 43.41% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved six nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. There was a total of 11549 positions in the final dataset. Evolutionary analyses were conducted in MEGA X.
Brycon nattereri voucher LAGEEVO_3928 mitochondrion, complete genome, Accession number MT428073.1: https://www.ncbi.nlm.nih.gov/nuccore/MT428073.1
Brycon nattereri voucher LAGEEVO_3847 mitochondrion, complete genome, Accession number MT428074.1: https://www.ncbi.nlm.nih.gov/nuccore/MT428074.1
Authors thank Henrique Peluzio for his support on the UFV cluster. S.V.R. thanks to CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) for the D.Sc. fellowship.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for creating the dataset(s) clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Morphological and molecular studies of parasitic helminths of freshwater fish and invertebrates (Davies) and Molecular and phylogenetic studies on parasites of clinical and ecological relevance (Lauthier).
Is the rationale for creating the dataset(s) clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genetics and molecular biology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 18 Nov 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)