Keywords
Dot Blot, FIJI, ImageJ macro, template registration, intensity measurements
This article is included in the NEUBIAS - the Bioimage Analysts Network gateway.
Dot Blot, FIJI, ImageJ macro, template registration, intensity measurements
A dot blot is a common technique in molecular biology to query the presence of an analyte in a solution. Commercially available dot blots contain a set of so-called capture antibodies spotted to a membrane in a given pattern, with each spot representing one specific antibody. Binding of the analyte to the capture antibody is detected by a detection antibody and chemiluminescence signals are recorded by film or scanner, similar as in classical western blots. Dot blots typically contain ~200 spots to be analyzed. For automatic quantification of the dot intensities a mask reflecting the spot pattern needs to be registered to the blot. Common registration algorithms are either intensity-based, as in e.g. StackReg1, or rely on the automatic identification of prominent features present in both images (e.g. Linear Stack Alignment with SIFT2)1,2. A scanned dot blot can often not be aligned to the mask with either method, especially when only a few analytes in the dot blot give positive signals. For algorithms that depend simply on feature detection, the pattern of the mask is too dense and regular to be successfully registered to few sparse dots. The intensity-based algorithm fails for similar reasons. As a result, open source tools currently available for analyzing dot blots either strongly rely on user input3 or assume full regular patterns4, which is not always the case. The presented ImageJ Macro DotBlot_Analysis.ijm relies on a few (3–5) landmarks selected by the user to give a first estimate of the transformation between the scanned dot blot and a template pattern. It then automatically searches for more landmarks and performs the final registration and measurements. It runs within the free and open-source software Fiji5.
The macro expects two images: the scanned dot blot and a mask image. The mask image is a representation of the spot pattern. After starting the macro, the user will be asked to select the names of the mask and dot blot images, as well as enter an expected spot size in pixel and a prominence value (discussed under Implementation). The user then needs to mark 3–5 corresponding dots (=landmarks) in the scanned dot blot and in the mask. DotBlot_Analysis.ijm uses the user-set landmarks to register the mask image to the scanned blot in an iterative process. After registration the macro creates selections for each spot. The user has the possibility to manually adjust the selections by activating a selection within the ROI manager, adjusting it and pressing “update” to save the new position. After this the macro performs a background subtraction and measures the intensities of each spot. The final output of the macro is a result table that lists intensity measurements for each spot. For quality control purposes, the macro also saves the selections within the ROI Manager as a zip-file and a control-image with all spot selections overlaid over the inverted background-subtracted dot blot. The zip-file can be re-opened in Fiji by drag&drop: all selections will be listed in the ROI Manager after re-opening.
The mask is a schematic representation of the spot pattern and needs to be registered to the scanned blot by translation, rotation and scaling (affine transformation). The macro registers the mask in two steps. In the first step it uses the 3–5 landmarks set by the user. The macro runs the plug-in “Landmark Correspondences”3 by Stephan Saalfeld, which matches the two sets of dots to determine the transformation and applies the transformation to the mask. In a second registration step, the macro automatically searches for more landmarks, in order to achieve an even finer registration. The landmarks are determined with the following computational steps:
1. Gaussian-filtering of the scanned, inverted dot blot. Signals are thereby smoothed and single signals are transformed to a smooth intensity distribution with (ideally) one single intensity maximum in the center of the spot.
2. Activation of the outlines of the single spots, as detected after the first registration step.
3. Search for local maxima on the gaussian-blurred image within each single spot outline. Maxima are determined as those pixels which are brighter than the local background, by the prominence value initially defined by the user. This ensures detection of bright signals and avoids eventual noise.
4. Once a local maximum is found, its position is marked as a new landmark. The corresponding landmark on the mask image is determined as the centroid of the selection that was used for searching the maximum.
Following this procedure, a new pair of landmarks (one set for the blot, one set for the mask) is identified and the plug-in “Landmark Correspondences” is re-run, now with more landmarks.
Once the mask has been fully registered to the scanned blot, it can be used for creating the final spot selections: the centroids of each spot of the final transformed mask are extracted and used to draw a new selection of defined size. This ensures that each spot is measured with a selection of equal size. The size of selection is provided by the user at the beginning of the script. The user has the possibility to correct the position of the final selections.
Before doing the measurements, the background of the image is subtracted using the “Subtract Background” command4, based on the rolling ball algorithm. The radius of the rolling ball is set to three times the user-defined spot size, to ensure that only background but not signal is removed.
After background subtraction the intensities of each spot are measured.
The final output of the macro is a result table that lists the following measurements for each spot:
- Label of the spot
- Area (for reporting, since it is the same area for all spots)
- Mean intensity (“Mean”)
- Minimum intensity (“Min”)
- Maximum intensity (“Max”)
- X- and Y-coordinate of the spot
- Intensity density: the product of area (scaled units, if set) and mean intensity
- RawIntDen: the sum of the pixel values within the spot
See the ImageJ documentation5 for more detailed information about the measurements.
The macro runs within the open-source and free software Fiji (https://fiji.sc/), which is supported on the following systems: Windows XP, Vista, 7, 8 and 10; Mac OS X 10.8 "Mountain Lion" or later; Linux on amd64 and x86 architectures.
The macro and the data for testing are available for download at https://github.com/ahklemm/AnalyzeDotBlots. Download the material from GitHub by clicking on the green “code” button and choose “download ZIP”. Unzip the material and open DotBlot_Analysis.ijm by drag&drop to Fiji. For testing purposes, open blot.tif and mask.tif. Run the macro.
Figure 1a shows as use case the scanned blot, blot.tif, and its mask, mask.tif. Four landmarks are marked in both the scanned blot and the mask (yellow arrows). Figure 1b shows the outcome after the first registration step. The blot has been inverted to create bright signals over dark background. While the spots already overlay the signals of the scanned dot blot quite well after this first registration, some selections (indicated with red arrow heads in Figure 1b) are offset with respect to the center of the actual signal. After the first registration the macro automatically identifies more landmarks, which are marked as yellow crosses in Figure 1c. The automatically identified landmarks are used for a second, finer registration step. The result of this second registration is visible in Figure 1d. Note that the center of the selections now overlays well with the center of the spots (red arrow heads, Figure 1d). Figure 1e shows the final result: the overlay of the final measurement selection over the background-corrected blot and the final results table.
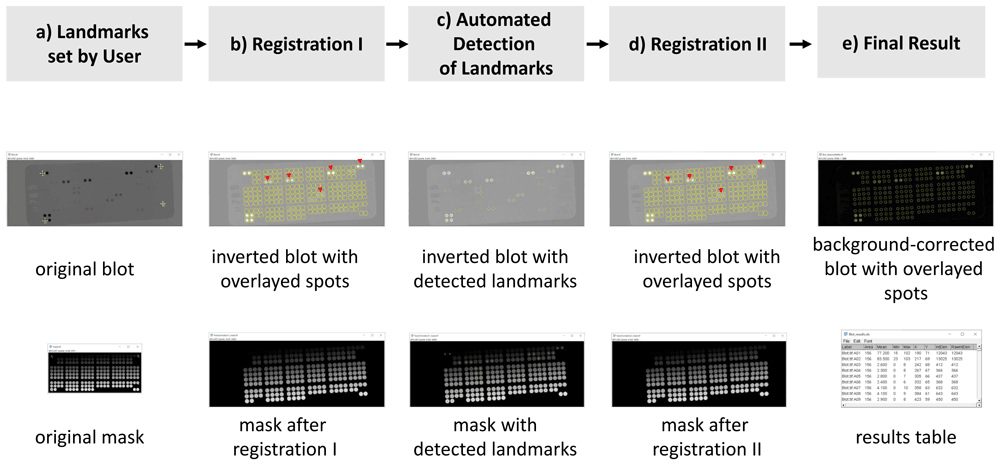
a) User selected landmarks in the scanned dot blot and the mask. b) Overlay of the spots over the dot blot after the first registration step. Red arrowheads mark spots with major shifts in respect to the signal center. c) Yellow crosses show the automatically detected landmarks in the scanned blot image as well as on the mask. d) Overlay of the spots after the second registration step. The marked spots (red arrowheads) now match the signal center better. e) Final results of the workflow.
The use case shows the scanned blot and mask of a very specific dot blot pattern/array. Unless the user wants to evaluate blots of exactly that array one needs to provide a new labelled mask. Within the labelled mask the spot on the upper left has an ID/gray-value = 1 and IDs/gray values of spots increase from the left to the right and from the top to the bottom. In mask.tif there are 218 spots, so the spot on the lower right has the gray value of 218. The ID/grayvalue is important in order to identify the spot even after the rotation/scaling/translation of the two affine registration steps.
MaskGenerator.ijm in the downloadable material helps to transform a typical vendor template6 into a labelled mask. Running MaskGenerator.ijm the user draws one selection around a spot of the template. The macro then identifies bright spots of the expected size and by roundness.
The presented Fijj macro DotBlot_Analysis.ijm supports users to analyze images of dot blots. It requires minimal input of few landmarks to then automatically measure the intensities of single spots in dot blots.
While scanning the dot blot one should avoid saturation of the signal. Signals should not reach a value of 0 but should always contain a minimum value of intensity. Reduce exposure times when imaging the film if needed.
Support for problems when using the script is provided via forum.image.sc – please link to the username of the author: @aklemm
Source code is available from: https://github.com/ahklemm/AnalyzeDotBlots
Archived source code as at time of publication: https://doi.org/10.5281/zenodo.41342196
License: BSD-3-Clause
The image of the dot blot was provided by Katja Eubler, Mayerhofer-lab, BMC, LMU Munich, Germany.
The author thanks Giovanni Cardone, MPI of Biochemistry, Munich for comments on the manuscript and the macro script and Andreas Thomae, CFBim, BMC, LMU Munich for discussions about the imaging of dot blots and analysis.
This publication was supported by COST Action NEUBIAS (CA15124), funded by COST (European Cooperation in Science and Technology).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: BioImage Analysis, Fluorescence Microscopy, Cell Biophysics
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bioimage analysis, computer vision, data science
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 01 Dec 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)