Keywords
myelin annotation tool, myelin quantification, fluorescence images, machine learning, image analysis
This article is included in the Artificial Intelligence and Machine Learning gateway.
This article is included in the NEUBIAS - the Bioimage Analysts Network gateway.
myelin annotation tool, myelin quantification, fluorescence images, machine learning, image analysis
The differences between CEM and CEMotates tools are clearly explained in the new version of the text. The advantages of CEMotate, which was developed by giving the time metrics of the tools and the precision differences of the experts, was indicated by numerical data.
See the authors' detailed response to the review by Predrag Janjic
Myelin degeneration causes neurodegenerative disorders, such as multiple sclerosis (MS)1,2. There are no remyelinating drugs. Myelin quantification is essential for drug discovery, which often involves screening thousands of compounds3. Currently, myelin quantification is manual, and labor-intensive. Automation of quantification using machine learning can facilitate drug discovery by reducing time and labor costs4. However, myelin annotation suffers the same limitations as manual quantification. To assist researchers and bioimage analysts, we developed a workflow and software for myelin ground truth extraction from multi-spectral fluorescent images.
Myelin is formed by oligodendrocytes wrapping the axons5. It is identified by continuous co-localization of cellular extensions that span multiple channels and z-sections (Figure 1). In our workflow, co-localizing pixels, candidate myelins, were determined using Computer-assisted Evaluation of Myelin (CEM) software that we previously developed6. In this context, CEM software functions as a candidate myelin detection program because it simply identifies overlapping pixels. Briefly, CEM removes cell bodies, defined as the overlap of nuclei and cellular marker, and identifies overlapping pixels between remaining oligodendrocyte and neuron channels6.
In the current study, the CEMotate tool7 was developed to efficiently evaluate these candidate myelins and to extract myelin ground truths. Using CEMotate, an RGB-composite z-section image, corresponding CEM output image, and expert’s markings can be visualized simultaneously to decide whether to keep or remove candidate pixels (see Implementation). The user can move along x-y-z axes and show/hide channels, images, and markings. Markings from the -1/+1 z-sections can be viewed simultaneously. Finally, CEMotate allows simultaneous visualization of myelin markings of two experts, which is important for inter-expert comparison.
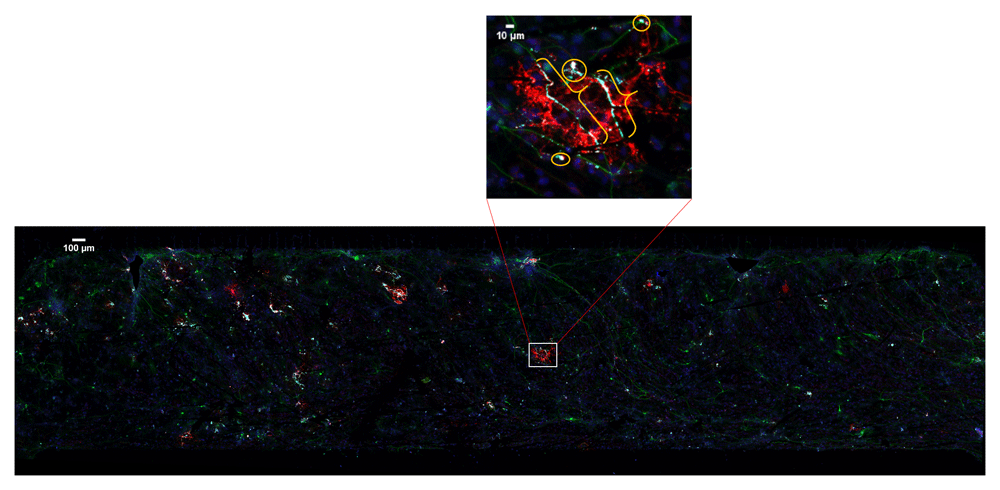
20× confocal microscopy image tiles were stitched together covering approximately 2 × 8 mm by 30–50 μm volume. Boxed area is enlarged to show myelin (brackets) and the false positive pixels (circles).
Using the described workflow, we annotated five images encompassing approximately 2 × 8 mm by 30–50 μm volume. The entire process, which would have taken several weeks, took approximately 5 days. More than 30,000 feature images were extracted from these five images and were used for testing various machine-learning methods8–10. The annotated images, which are available with the manuscript, are a resource for the researchers working not only on myelin detection but also on segmenting multi-spectral images.
Images were previously acquired6. Briefly, co-cultures of mouse embryonic stem cell-derived oligodendrocytes and neurons were grown in microfluidic chambers. After myelin formation, cells were fixed in paraformaldehyde and were stained with 1:1,000 mouse or rabbit anti-TUJ1 (Covance), 1:50 rat anti-MBP (Serotec), and DAPI (Sigma). Images were acquired on Zeiss confocal microscopes as tiles approximately 2mm×8mm. The z-axis, 30–50 µm, was covered by 1-µm-thick optical z-sections. The tiles were stitched together on Zen software (Zeiss). No further processing was done.
In CEMotate, a new project is started by loading oligodendrocyte, axon, and nucleus images, red, green, and blue channels respectively in the example (Figure 2). Users can save and reopen projects. In CEMotate, users can zoom using the mouse wheel and can move in the x-y axes and z-axis using scroll bars and buttons respectively (Figure 2 and Figure 3).
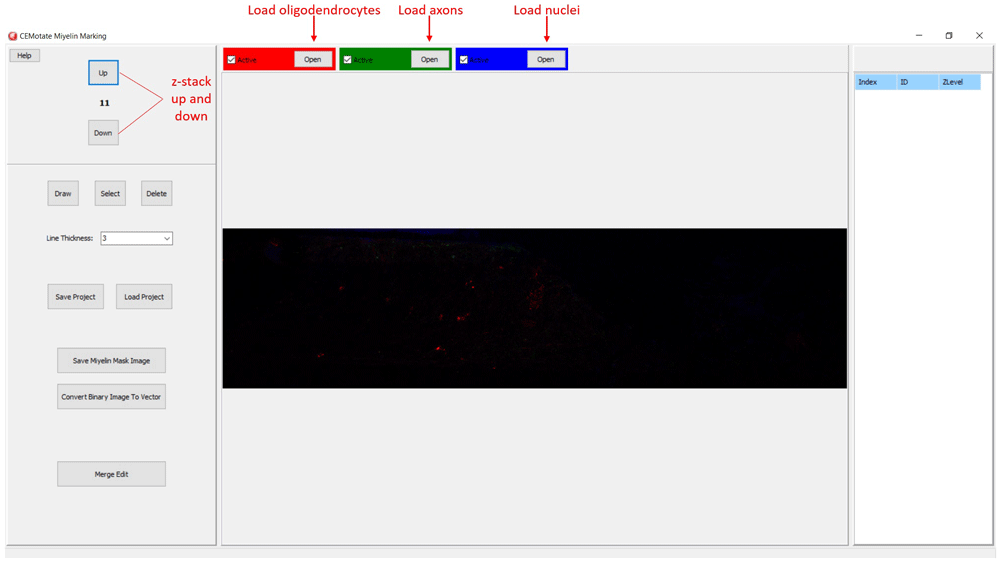
Buttons for loading oligodendrocyte, axon, and nucleus images, and navigating the z-stack button to up and down are marked.
Myelin pixels may be marked at various thickness values (Figure 3). CEMotate records myelin drawings as vectors in the “.iev” files. These vectors can be modified or deleted in CEMotate (Figure 3). Optionally, to facilitate myelin detection, the candidate myelins can be loaded from CEM6 or another source that generates binary images of myelin markings. Myelin identification using CEM is described in detail in 6. Output of CEM, is a binary image, which is converted to vectors using the included module (Figure 4). Note that the conversion will overwrite your existing myelin vectors.
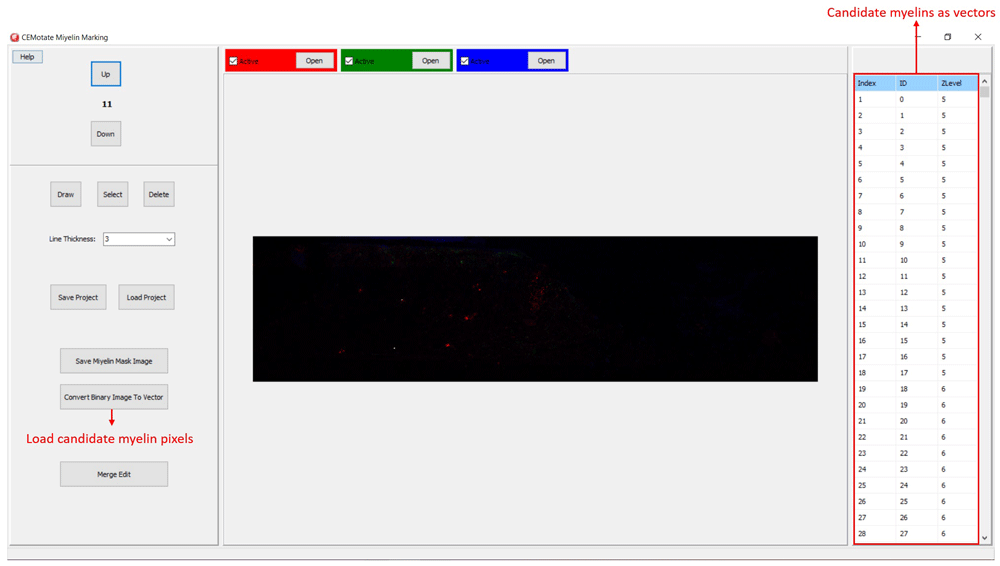
To load candidate myelin pixels, use “Convert Binary Image to Vector” button.
Additionally, myelin regions from two sources can be visualized simultaneously. This allows visualization of myelins annotated by experts and CEM, to do so, first, rename and copy the ‘‘.iev’’ file containing second myelin vectors to the same folder. Next, modify the ‘‘.ini” files as shown in Figure 5. After loading the modified ‘‘.ini” file using the ‘Merge Edit’ button, myelin vectors will be shown in two different colors (Figure 6). These vectors can be modified as in Figure 6.
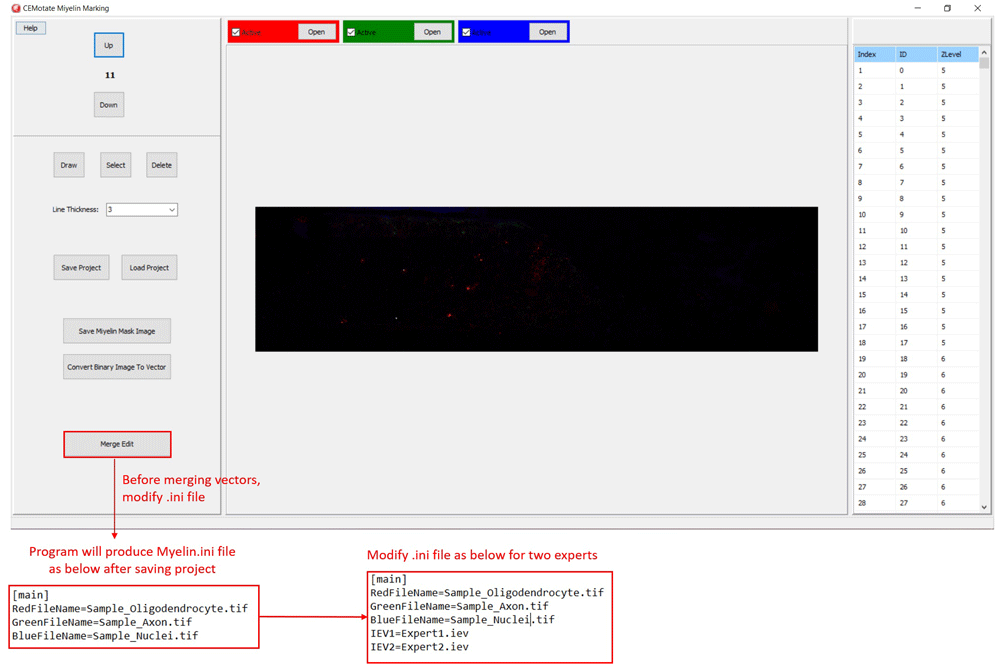
Modify .ini file as in the lower panels and load it using “Merge Edit” button.
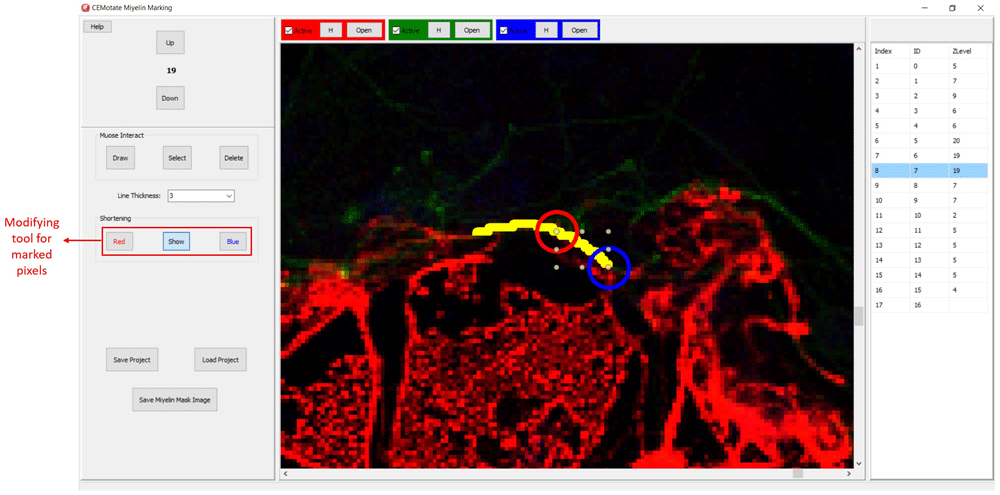
CEM candidate myelins or two experts’ markings can be shortened, deleted or drawn over.
Once done with marking, users can convert the myelin vectors into an image using the “Save Myelin Mask Image” button. We implemented this strategy to extract gold standard myelin ground truths.
The myelins marked by two experts were compared against the gold standards. Experts’ precision for each image was calculated as described in 9. The average precision was calculated as mean of precision values of each expert for each image.
In this study, myelin was marked by two experts on previously acquired oligodendrocyte and neuron co-culture images6 using the described workflow (see Implementation). A third expert evaluated their markings and extracted gold standard myelin ground truths. The ground truth images were saved as TIF on CEMotate7. All images are available (see below).
Each image covered a large volume (approximately 2 × 8 mm by 30–50 μm).
While CEM determined the candidate myelins on five images in approximately 43 minutes, ML approach took only 1.04 seconds for the same process8 (Table 1). Extracting the gold standard myelin ground truths from five images with candidate pixels that were determined by CEM took approximately another 35 hours for one expert. This process involved determining FPs and FNs on ImageJ. The same process took approximately 20 hours for an expert using CEMotate. Thus, over 40% of time was saved (Table 2). Moreover, CEMotate enabled collaboration of three experts for accelerated myelin ground truth extraction. Because ImageJ does not have such a feature, we could not directly compare the times saved for this process.
| CEM | ML Approach9 | |
|---|---|---|
| Time (~) | 43 min | 1.04 sec |
| Expert 1 | Expert 2 | |
|---|---|---|
| Average Precisions | 36.23% | 60.54% |
CEM identified 219032 candidate myelin pixels on five images. Two experts identified TP myelins. A third expert evaluated these results to obtain the gold standard myelin ground truths which covered 9550 pixels. To the best of our knowledge, this is the first time myelin ground truths are shared with the science community.
Next, we calculated each expert's performance (Table 3). Two experts averaged 48.39% precision. The highest precision of an expert was 87.95% for one image. In comparison, our customized-CNN and Boosted Trees consistently reached precision values over 99%8. These results suggest that, machine learning methods can outperform human annotators once trained with accurately labeled data.
CEMotate7 accelerates annotation of multi-spectral images. As an example, we used it to annotate myelin, which can only be identified as co-localization of neuron and oligodendrocyte membranes within certain criteria. CEMotate’s visualization features simplified inter-expert collaboration and validation. Moreover, myelin ground truths accompanying this manuscript are a resource for the researchers working on segmenting myelin and other features in multi-spectral images.
Image Data Resource: A Multi-Spectral Myelin Annotation Tool for Machine Learning Based Myelin Quantification. Project number idr0100; https://doi.org/10.17867/1000015211.
This project contains the raw image files analyzed in this article.
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
CEM and CEMotate are available from: https://github.com/ArgenitTech/Neubias.
Archived source code as at the time of publication: https://doi.org/10.5281/zenodo.41083217.
License: Non-Profit Open Software License 3.0 (NPOSL-3.0).
This publication was supported by COST Action NEUBIAS (CA15124), funded by COST (European Cooperation in Science and Technology).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Computational neuroscience, structural studies of white matter, dynamical models of glial membrane.
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
No
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
No
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: computational neuroscience, structural studies of white matter, dynamical models of glial membrane.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 4 (revision) 15 Nov 23 |
read | |
|
Version 3 (revision) 27 Apr 22 |
read | read |
|
Version 2 (revision) 09 Mar 22 |
read | |
|
Version 1 21 Dec 20 |
read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)