Keywords
anthocyanins, microRNAs, fruit development, RNA interference, evolution
anthocyanins, microRNAs, fruit development, RNA interference, evolution
Two citations (20, 21) were added as suggested by Reviewer 2 to document prior uses of sRNA libraries as confirmatory evidence of secondary sRNAs as products of miRNA targeting. A software analysis was added as suggested by Reviewer 2: “strucVis” for visualization of predicted RNA secondary structures with overlaid sRNA depths display of ShortStack output. Extended data: Figure S1 was revised to include the strucVis analysis and the legend modified as requested by Reviewer 2. The titles of Extended data: Tables S1 and S2 were changed for clarity as requested by Reviewer 1. The legend to Figure 1 was modified as requested by Reviewer 2 to describe the images as T plots of 5’ ends of sRNA alignments, which I previously (citation 7) named as pseudo-degradome analyses and referenced in the software availability description. The legend of Figure 2 was modified to direct readers to Extended data Tables where underlying evidence is documented and file rows are referenced to substantiate claims, as requested by Reviewer 2. Minor changes to the text were made as requested by Reviewers to improve clarity and articulate comparisons made across Figure 2 panels for interpretations underlying claims. Reviewer 2’s suggestion to elaborate on methods was addressed by a statement added at the end of the Software availability section: “The options parameters used for various algorithms are detailed in Extended data: Tables S1- S3.” No changes were made to data in Figures or Extended data Tables in this revision.
See the author's detailed response to the review by Brian D. Gregory
See the author's detailed response to the review by Nathan Johnson
Chen et al.1 sequenced Ananas comosus var. bracteatus accession CB5, cultivated for its bright pink-to-red colored fruit, and yellow-fleshed A. comosus accession F153, reporting an improved F153 reference assembly2 while annotating MICRORNA (MIRNA) loci2–4 and gene family expressions relevant to lignin and anthocyanin biosynthesis. An independent article reported var. bracteatus MIRNAs5 but not MIR8286, a negative regulator of anthocyanin and polyphenolics biosynthesis by targeting MYB transcription factors associated with UV light- and sugar-signaling in dicots7,8. MIR828 has been reported in gymnosperms8, Amborella (sister to flowering plants)9, and basal monocot orders Liliales8,10, Asparagales11, Zingiberales12,13, Arecales14, but not in the Poales, a sister order comprising grasses and ~3,000 species of bromeliads including pineapple15. Here I show MIR828 exists in pineapple and directs post-transcriptional gene silencing of mRNAs encoding MYB family members with inferred function to regulate the conspicuous red fruit trait in var. bracteatus. MIR828 plesiomorphy (an ancient basal trait) may shed light on monocot apomorphic fruit development, postulated for 21 monocot families with fleshy fruits as due to homoplasy/convergence driven by tropical climate and/or enticements to vertebrate endozoic seed dispersers16.
The astronomer Carl Sagan popularized the quip (a corollary of Occam’s Razor) “absence of evidence is NOT evidence of absence.” Taking a conservative approach applies especially to quantitative transcriptomics. For example, the existence of regulatory and post-transcriptional processes increase the complexity of non-coding RNA space, where annotation is sparse. Analysis of Chen et al’s A. comosus MD-2 cultivar leaf small RNA (sRNA) libraries and stranded RNA-seq libraries4 from flowers and fruits of F153 and CB5 genotypes establish the existence of aco-miR828 (Extended data: Figure S117) and pri-MIR828 expressions, with the novel observation that pri-MIR828 is properly transcribed in leaves, flowers and fruits yet ~50% of mature miR828 species abundance (0.3 reads per million in leaves from 264.7 million reads; Extended data: Table S117) are 21 nt, while 93% of the equally abundant 22 nt species appear to have undergone non-templated 3’ uridylation of the 21 nt species18 (Extended data: Table S117). Analysis of independent bracteatus cultivar leaf sRNA libraries (NCBI SRA SRR5677552-7; 113.2 million reads)5 of unknown provenance relative to the subject CB5 genotype failed to identify any MIR828 reads.
Fortuitously, a unique aspect of miR828 is that despite its very low abundance, it has very high activity7,19 that serves as diagnostic. miR828 guides ARGONAUTE slicing of target MYB mRNAs within the deeply conserved SANT domain region8 by Watson-Crick complementarity, with consequent knock-on production of easily quantified DICER-mediated sense- and antisense 21 nt phased small-interfering RNAs (phasiRNAs) mapping downstream (3’) on target MYB transcripts. The improved F153 reference assembly1 contains two candidate miR828-targeted MYBs: Aco017254.1 (LG4), with two introns of 1113 and 1340 nts, and Aco020986.1 (LG14) without RNA-seq evidence of intron splicing, whereas the CB5 bracteatus genome only contains one Aco017254.1 homologous gene with RNA-seq splicing evidence for conserved introns of 1109 and 1332 nt (Extended data: Table S217). When phasiRNA expressions from leaf sRNA libraries4,5 are respectively mapped to the F153 and CB5 candidate MYB target mRNAs, it is apparent that F153 MYB transcripts clearly undergo miR828-guided slicing, evidenced by D1(+) phased siRNA reads mapping to the 10th nucleotide position of miR828 homology to the mRNA target, and unique sense and antisense secondary phasi-RNAs mapping precisely in multiples of 21 nt downstream from the detected slice sites (Figure 1; Extended data: Figure S2, Table S317). The CB5 reference genome target MYB locus Aco017254.1 homolog (contig tig00012294, CABWKS010000088.1:25590-28868rc) encodes two missense codons at residues 41 (M➔R) and 200 (N➔K compared to F153), four silent codon substitutions, and RNA-seq analysis reveals mis-annotation of the CB5 genome which lacks two Gs (at contig residues 28780 and 27566) that exist and result in a CB5 open reading frame of the same size as Aco017254.1 in F153, including six instead of seven trinucleotide GGC glycine codon repeats templated in the CB5 genome at residue 204 (Extended data: Table S217).
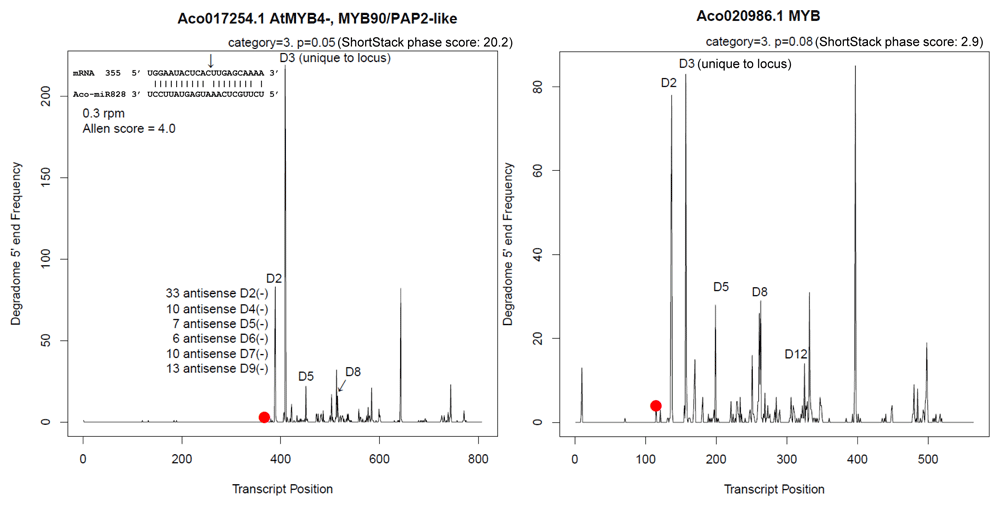
Concatenated sRNA libraries were used as pseudo-degradome on grounds sliced mRNAs subject to production of amplified diced dsRNAs are manifest in sRNA libraries7,20,21. Slicing is at nt10 from 5' end of miR828 (arrow, inset; same target sequence for Aco020986.1 at mRNA nt 115). Y axis is numbers of reads in sRNA libraries mapped to cDNAs; red dot is the documented miR828 sliced sRNA species that sets the register for 3’ phasiRNA production.
In contrast to demonstrated post-transcriptional silencing activity of miR828 on target MYB abundance in F153 leaf samples (Figure 1), analysis of six bracteatus leaf sRNA libraries from an independent study5 did not provide any evidence of target MYB Aco017254.1 mRNA slicing or target phasiRNA accumulation (Extended data: Table S317). Taken together, subject to the caveat that the provenance of the bracteatus cultivar used for the sRNA analysis5 may be different than subject CB5 genotype, the data suggest there may be differences in expression and/or regulation of pri-MIR828, and/or target MYB Aco017254.1 between F153 yellow-fleshed versus CB5 red-fleshed genotypes. Figure 2 shows this is indeed the case in various tissues examined, with the evidence supporting higher pri-MIR828 expression in F153 ovules concordant with lower target MYB Aco017254.1 mRNA levels (Figure 2A), significant decreases over time from stage 1 early fruit development for target MYBs Aco017254.1 and Aco020986.1 in yellow-fleshed MD-2 cultivar (Figure 2B), whereas in contrast there is a trend of lower expression of CB5 pri-MIR828 concordant with sustained higher abundance of Aco017254.1 target MYB (Figure 2C stage 7 versus stage 1) than seen in MD-2 during the ripening stage of red-fleshed CB5 genotype (compare Figure 2C stage 7 showing high CB5 Aco017254.1 abundance to Figure 2B stage 7 showing very low MD-2 Aco017254.1 abundance).
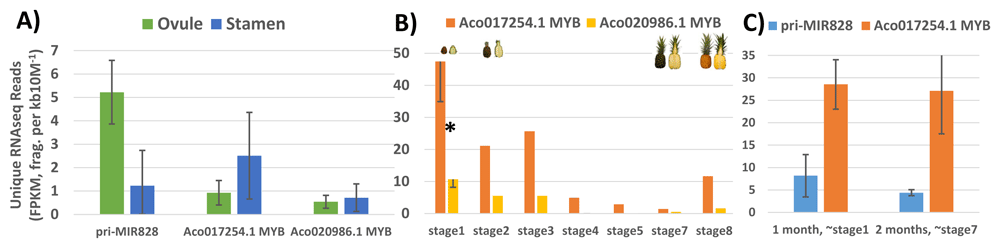
A) Inverse relationship of pri-MIR828 (high; see Extended data: Table S1, rows 23–36) to target MYB Aco017254.1 (low Extended data: Table S2, rows 406–470) anthocyanin effector expressions in F153 ovules. Error bars are s.e.m., n=3 biological replicates. B) Down regulation of miR828-targeted MYB anthocyanin effectors during MD2 yellow-fleshed fruit development (n=1 per stage Extended data: Table S2, rows 134–405)). Asterisk (*) denotes stage 1 significantly different across Aco017254.1 and Aco020986.1 than stages 2-8, p = 0.02 (Student’s two-sided paired t-test, equal variance assumed). Stage 1 error bars show the 95% confidence interval for significance. C) Apparent maintenance of positive anthocyanin effector MYB Aco017254.1 expression at “ripe” two-month stage of CB5 red-fleshed fruit development, concordant with trend of lower pri-MIR828 expression (see Extended data: Table S1, rows 37–52; Table S2, rows 471–883). Error bars are s.e.m., n=5 biological replicates.
Plant development gives rise to an astounding complexity of shapes, colors, and functions that Darwin called ‘an abominable mystery’ in his efforts to integrate species complexity with the unifying theory of evolution. The observations reported here potentially offer insight into the conservation of a developmental control mechanism whereby miR828 in pineapple, like in dicots silences MYBs inferred to act as positive effectors of anthocyanin biosynthesis that could give rise to the red-fleshed trait in the bracteatus variety. Consistent with this view is the finding from genome-wide functional phylogenomic approaches that ARGONAUTE1, RNA-DEPENDENT RNA POLYMERASE6, and mRNA export factor homolog SILENCING DEFECTIVE5, each required for trans-acting siRNA accumulation, played significant roles in the evolution of monocot metabolic and developmental traits22,23. Also of interest in this context is the claim of bracteatus genome authors24 that convergent expansion in several Crassulacean Acid Metabolism (CAM) bromeliad lineages of XAP5 CIRCADIAN TIMEKEEPER/XCT, a nuclear-localized regulator of blue light responses25 and sRNA production26, supports evolution of the myriad metabolic and physiological transitions from C3 to CAM photosynthesis by duplication/differentiation of a highly pleiotropic effector24.
RNA-seq data1, CB5 reference genome GCA_902506285.1, and F153 improved assembly genome GCA_902162155.1 were downloaded from NCBI via BioProject accession code PRJEB33121. sRNA raw data5 for A. comosus var. bracteatus leaf libraries were downloaded from NCBI BioProject accession code PRJNA389361. Processed data4 for MD-2 leaf sRNA libraries Pn_Gr2am_1, Pn_Gr3pm_3, Pn_Gr4pm_1, Pn_Gr4pm_3, Pn_Gr6am_2, Pn_GrMid_3, Pn_Wh10am_2, Pn_Wh10pm_1, Pn_Wh1pm_1, Pn_Wh2am_1, Pn_Wh2am_2, Pn_Wh3pm_1, Pn_Wh3pm_2, Pn_Wh4am_1, Pn_Wh4pm_1, Pn_Wh6am_1, Pn_Wh6am_2, Pn_Wh6pm_1, Pn_Wh6pm_2, Pn_Wh8am_2, Pn_Wh8pm_2, Pn_WhMid_1, and Pn_WhMid_2 were downloaded from https://mpss.danforthcenter.org/dbs/index.php?SITE=pineapple_sRNA.
The original A. comosus F153 reference genome and cDNA fasta file ‘Acomosus_321_v3.cds.fa’ can be browsed online and downloaded at https://phytozome.jgi.doe.gov/27.
Figshare: A role for MIR828 in pineapple fruit development, https://doi.org/10.6084/m9.figshare.11388051.v217
This project contains the following extended data:
Figure S1. strucVis graphical linear output of Aco-MIR828 hairpin structure and sRNA abundance evidence from 23 MD-2 leaf sRNA libraries.
Figure S2. PhaseTank alignment output of leaf MD-2 sRNA libraries mapped to candidate miR828 target MYB cDNAs, F153 improved reference genome.
Table S1: Evidences for pri-MIR828 abundance in F153 and CB5 varieties.
Table S2: RNA-seq evidence for miR828 target MYB abundance.
Table S3: bowtie mapping of sRNAs to miR828 target MYB mRNAs
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Fastx-toolkit version 0.0.14 for trimming Illumina universal adapterAGATCGGAAGAGCACACGTCTGAACTCCAGTCA (fastx_clipper option -l 18; discards short reads) and fasta file manipulations (fastx_uncollapser to expand the pre-processed data4; fastq_to_fasta for inputs to CleaveLand) is available at http://hannonlab.cshl.edu/fastx_toolkit/. FastQC version 0.11.5 for quality control of fastq raw sequence data is available at https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. ShortStack28 version 3.8.5 (options --mincov 15 --foldsize 340; Extended data: Figure S1, Table S117) for comprehensive annotation and quantitation of MIRNAs and sRNA cluster phasing is available at https://github.com/MikeAxtell/ShortStack. strucVis for visualization of predicted RNA secondary structures with overlaid sRNA depths display of ShortStack output (Extended data: Figure S1, page 2) is available at https://github.com/MikeAxtell/strucVis. CleaveLand29 version 4.4 for plotting 5’ ends of sRNA libraries as pseudo-degradome inputs (amplified sense and anti-sense siRNAs in phase and derived from miRNA-sliced mRNAs7,20,21) to find sliced miRNA targets is available at https://github.com/MikeAxtell/CleaveLand4. PhaseTank30 version 1.0 for quantifying and aligning phasiRNAs to miRNA target mRNAs (Extended data: Figure S217) is available at http://phasetank.sourceforge.net/; the linux command sed -i ‘s/-/_x/’ was used to reconfigure fastx_collapser output from concatenated sRNA fasta library files to PhaseTank input style “>t1_xN” where N is number of collapsed reads). Magic-BLAST31 version 1.5.0 for RNA-seq fastq read alignment (Extended data: Table S2 row 4 for options parameters17) to reference genomes is available at https://ncbi.github.io/magicblast/. Blastn32 version 2.6.0 is available at ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/. Bowtie33 version 1.1.2 for short read alignment (option -v 1 to allow one mismatch) is available at http://bowtie-bio.sourceforge.net/manual.shtml. RNAfold34 web server was used for generating miR828 hairpin graphics (Extended data: Figure S117) at http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi. The options parameters used for various algorithms are detailed in Extended data: Tables S1–S3
The author thanks the TTU High Performance Computer Center for support in use of the Quanah supercluster, and Sunitha Sukumaran for help with perl scripting.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Small RNA analysis and annotation in plants.
Is the rationale for commenting on the previous publication clearly described?
Yes
Are any opinions stated well-argued, clear and cogent?
Yes
Are arguments sufficiently supported by evidence from the published literature or by new data and results?
Partly
Is the conclusion balanced and justified on the basis of the presented arguments?
Partly
References
1. Shahid S, Kim G, Johnson N, Wafula E, et al.: MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature. 2018; 553 (7686): 82-85 Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Small RNA analysis and annotation in plants.
Is the rationale for commenting on the previous publication clearly described?
Yes
Are any opinions stated well-argued, clear and cogent?
Yes
Are arguments sufficiently supported by evidence from the published literature or by new data and results?
Yes
Is the conclusion balanced and justified on the basis of the presented arguments?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Plant RNA biology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 08 Apr 20 |
read | |
|
Version 1 13 Jan 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)