Keywords
Metallothionein, ccRCC, TCGA, Oncomine,Bioinformatics
This article is included in the Oncology gateway.
Metallothionein, ccRCC, TCGA, Oncomine,Bioinformatics
Clear cell renal cell carcinoma (ccRCC) is the most common type of renal tumor, which approximately constitutes 3% of adult malignancies and accounts for 90% of all renal malignancies1,2. In recent years, the understanding of the genetics and biology of RCC had been improved dramatically, however, the mortality for patients with advanced or metastatic ccRCC is still high3–5. Therefore, it is essential to find new biomarkers to improve the diagnosis and prognosis of ccRCC patients.
Metallothioneins (MTs) are a family of cysteine-rich proteins. Their molecular weight ranges from 6 to 7 kDa and they play essential roles in tumor biology such as proliferation, differentiation, apoptosis, and drug-resistance6–11. MT proteins are encoded by MT1-4 genes family located on chromosome 16q1312, among them, MT2-MT4 are encoded by a single gene, while MT1 isoforms were encoded by a set of 14 genes, which consists of eight protein coding genes (MT-1A, MT-1B, MT-1E, MT-1F, MT-1G, MT-1H, MT-1M and MT-1X) and six pseudogenes (MT-1C, MT-1D, MT-1I, MT-1J, MT-1K and MT-1L)13–15. The expression of MT1 isoforms vary at cellular levels and in different tissues13. Changes in MT1 isoform expression were detected in various types of human malignancies14–18. Therefore, MT1 isoform profiling had been utilized as diagnostic markers and as prognostic predictors of tumor diseases12–14. Lots of work has been performed regarding the expression and prognostic value of MT1 isoforms in ccRCC19–22. However, there was still not enough information about the expression and prognostic value of the MT1 isoforms in ccRCC. Thus, it would be of great importance to elucidate the pattern of MT1 isoform expression and evaluate their prognostic value in ccRCC for patients' benefits.
The present study aimed to explore mRNA expression status of MT1 isoforms and their prognostic significance in ccRCC. For this purpose, we firstly investigated mRNA expression levels of MT1 isoforms in human ccRCC using The Cancer Genome Atlas (TCGA), Gene Expression Profiling and Interactive Analysis (GEPIA) and Oncomine database. Secondly, prognostic value of MT1 isoforms mRNA expression in ccRCC patients was evaluated through GEPIA by analyzing clinical data from the TCGA-KIRC project. Finally, correlations of MT1G with other MT1 isoforms were conducted using the R program to show the correlation in ccRCC.
TCGA23 is a publicly available and open access platform. As a result, ethics committee approval was not required for use of the data from this platform. GEPIA (Peking University, Peking, China) was used to assess the association between gene expression and prognosis24,25. GEPIA adopts a standard processing pipeline to analyze the RNA-Seq expression data from GTEx and TCGA, which include 8,587 normal and 9,736 tumor samples. Here, we selected the MT1 family genes, including, MT1G, MT1F, MT1X, MT1H, MT1A, MT1E, MT1M, MT1L, and the cancer types (Kidney Renal Clear Cell Carcinoma; KIRC). For the gene mRNA level expression between normal and tumor tissue in ccRCC, a boxplot using disease state as a variable was graphed to calculate differential expression of MT1 isoforms. We obtained samples from TCGA and used GEPIA to analyze the connections between overall survival and disease-free survival rates with MT1 isoforms expression in KIRC. Log-rank test was used to test the difference between Kaplan-Meier survival curves. Kaplan-Meier survival analysis was performed to analyze the relationship between the expression of MT1 isoforms and the survival days of KIRC patients25. MT1s were entered into the database to obtain Kaplan-Meier survival plots. The log rank p values and hazard ratios were calculated. A p-value < 0.05 was considered statistically significant.
Analysis of MT1 isoform expression in ccRCC was performed using the Oncomine database (www.oncomine.org; Compendia Biosciences, Ann Arbor, MI, USA). Oncomine can be accessed through the Oncomine Research Edition using an institution supplied email address26. We compared expression of the MT1 isoforms in clinical specimens of tumor datasets that contain both normal and tumor tissue. We further analyzed the MTIG gene expression in the six projects which contain both normal and tumor tissues of ccRCC. The six projects were Higgins Renal, Cutcliffe Renal, Gumz Renal, Jones Renal, Beroukhim Renal, Yusenko Renal and Lenburg Renal. Data were extracted to evaluate the expression of MT1 isoforms in RCC. In the study, p = 0.05, a fold change of 2, and a gene rank in the top 10% were set as the significance thresholds. Student's t test was used to analyze the difference in the expression of MT1s in RCC.
Gene expression in KIRC from the TCGA dataset was obtained through the ‘cgdsr’ R package (version 3.6.2). The mRNA sequence data and corresponding clinical traits of the TCGA-KIRC project were downloaded from TCGA, which contained 537 ccRCC tumor tissues. Gene symbol annotation information was used to match probes with corresponding genes. The correlation of MT1G with other MT1 isoforms was calculated by cor function in R using Spearman correlation and visualized using ggplot2 (version 3.3.0) and ggpubr (version 0.2.5) packages in R (version 3.6.2) in R-Studio (version 1.2.5033). The code is supplied as Underlying data.
To evaluate the MT1 isoforms mRNA expression levels in ccRCC normal and tumor tissues, we firstly analyzed transcriptome data from the TCGA-KIRC project using GEPIA. We found that mRNA expression levels of six MT1 isoforms, including MT1G, MT1H, MT1F, MT1X, MT1E and MT1M were simultaneously decreased in ccRCC tumor tissues compared with normal tissues (Figure 1). The results indicated that six out of eight transcripts of MT1 isoforms were significantly downregulated in ccRCC.
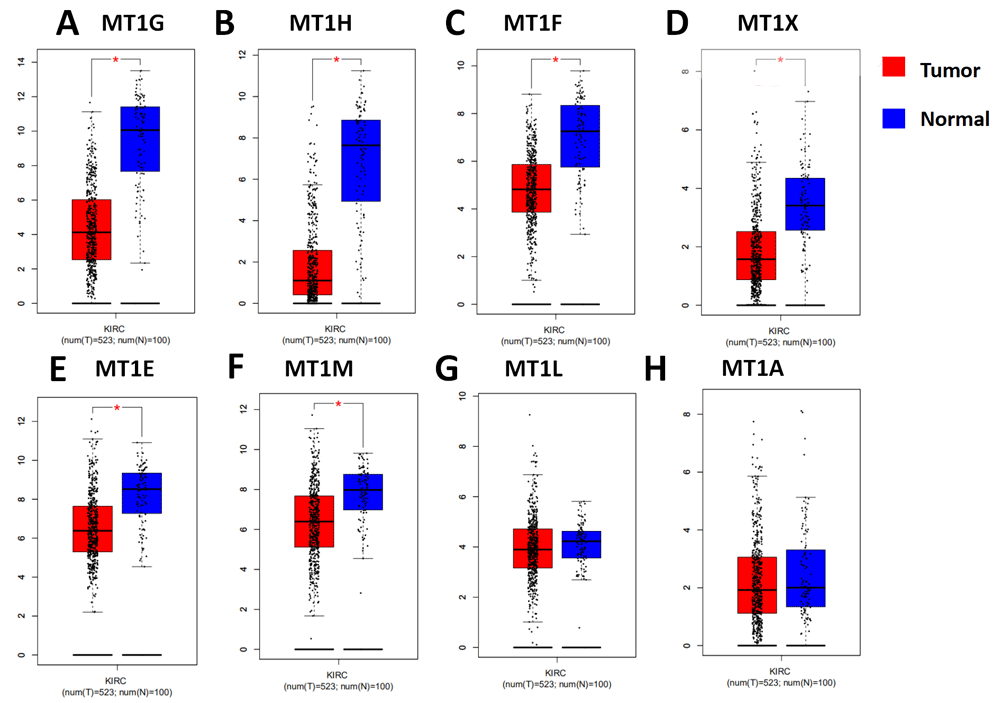
(A) MT1G, (B) MT1H, (C) MT1F, (D) MT1X, (E) MT1E, (F) MT1M, (G) MT1L and (H) MT1A. The criteria thresholds for datasets were logFC >1; *p<0.01.
To further confirm the expression levels of MT1 isoform mRNA in clinical samples, we screened the Oncomine database to investigate mRNA expression of MT1 isoforms between tumor and normal tissues. As shown in Figure 2A, eight MT1 isoform mRNA levels decreased in tumor tissues compared to normal tissues. In renal tumor cases, decreased mRNA expression of MT1 isoforms were detected in a total of nine datasets for MT1G, seven for MT1F, five for MT1H and MT1X, four for MT1H, four for MT1E, and two for MT1M. Taken together, these results indicate that mRNA expression levels of MT1 isoforms are downregulated in renal tumor.
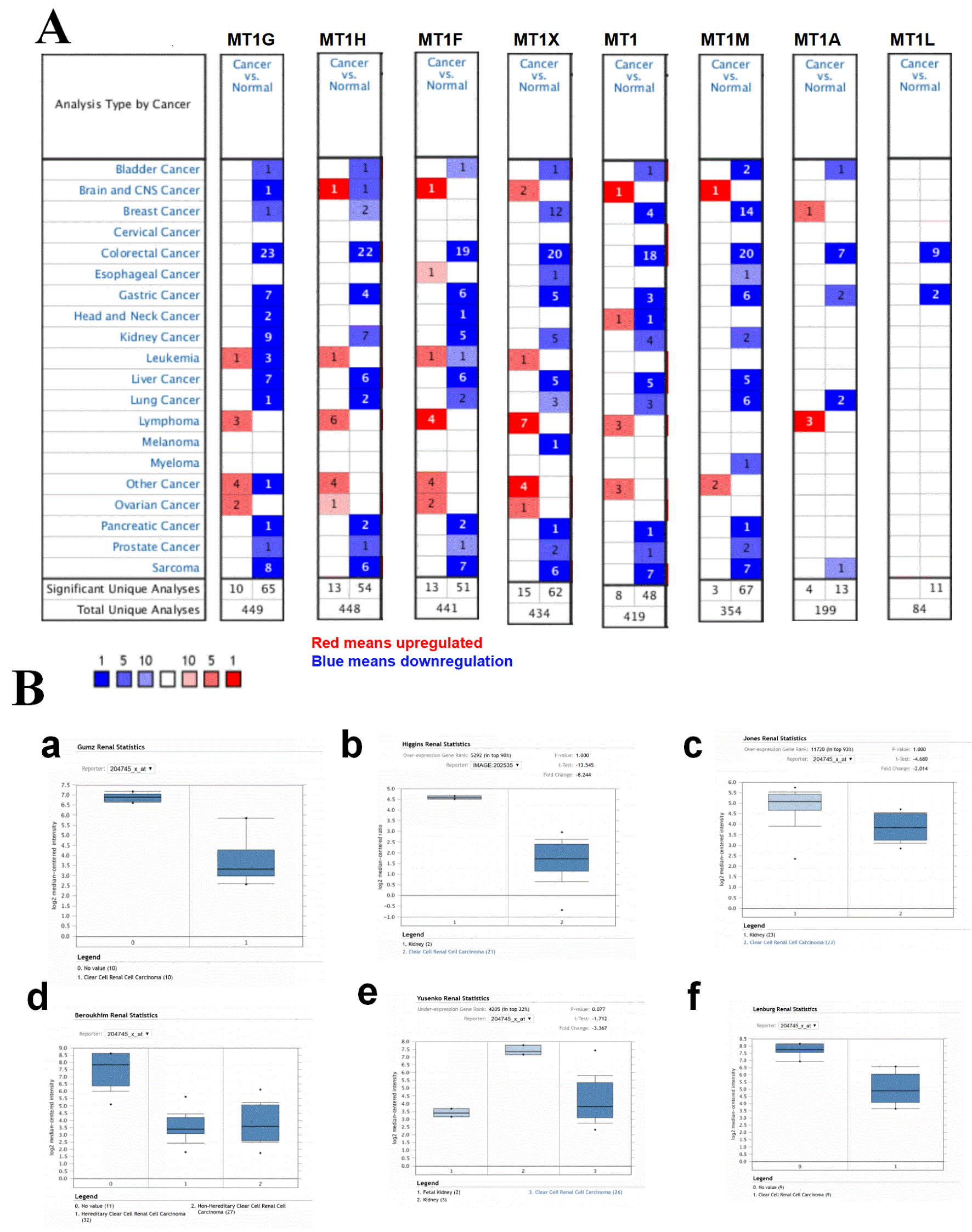
(A) Expressions of MT1 isoform mRNA in 20 common cancers were compared with those in corresponding normal tissues. The search criteria thresholds for datasets of cancer versus normal analysis were p-value of <0.0001, a fold change of >2, and a gene rank in the top 10%. Red signifies gene upregulated in the analyses; blue represents gene downregulated. (B) Confirmation of the MT1G decreased expression in clear cell renal cell carcinoma tumor tissues compared to normal tissues based on the Oncomine database in different projects.
To investigate the decreased expression of MT1G mRNA expression level in ccRCC tissues, we investigated the differences in mRNA level of MT1G between tumor and normal tissues in ccRCC using the Oncomine database. As shown in Figure 2B, there were six ccRCC projects containing tumor and normal tissues and all the datasets indicated that the expression levels of MT1G dramatically decreased in tumor tissues. Taken together, these results from clinical specimens demonstrate that mRNA expression levels of MT1G are significantly downregulated in ccRCC tumor tissues.
To figure out the MT1 isoform transcripts that are co-expressed with the MT1G transcript, we performed a correlation analysis between mRNA expression levels of MT1G and other MT1 isoform transcripts based on TCGA-KIRC transcriptome data (Spearman correlation shown in Figure 3). A strong correlation of mRNA expression in cancer tissues (r≥0.8) was found between MT1G and MT1H, MT1G and MT1F, a moderate correlation (r≥0.5) existed between MT1G and MT1M, MT1X or MT1E. The expression of eight MT1 isoform mRNA and clinicopathological information involved in this study are supplied in Underlying data.
To assess the value of eight MT1 isoforms in ccRCC prognosis, the relevance between MT1 isoforms and overall survival time and disease-free survival time were investigated. First, we examined the prognostic relevance of eight MT1 isoform mRNA expression in ccRCC using GEPIA survival analysis based on TCGA-KIRC clinicopathological data. The patients from the TCGA-KIRC dataset (n = 516, which have both the transcriptome data and clinicopathological data) were classified into low- and high-expression groups according to the median of the specific gene mRNA expression level. For MT1G, the clinicopathological information of different MT1G expression groups is shown in Table 1; survival differences between the low and high expression groups were detected with Kaplan-Meier survival curves. mRNA expression levels of MT1G, MT1H, MT1F and MT1X were inversely related to survival according to the Kaplan-Meier survival analysis (Figure 4). However, mRNA expression levels of MT1A, MT1E, MT1L, and MT1M were not significantly associated with the clinical outcome of ccRCC patients. We also found a shorter disease-free survival time in patients with higher mRNA expression of MT1G, MT1F and MT1X (Figure 5). To answer whether mRNA expression levels of MT1 isoforms correlated with ccRCC progression, we analyzed the relationship between MT1 isoform mRNA expression levels and tumor stage. As Figure 6 shows, MT1F and MT1X mRNA expression levels were positively associated with tumor stage. Collectively, these results indicate that high mRNA expression levels of MT1G, MT1F, and MT1X are associated with poor prognosis in ccRCC patients and that MT1F and MT1X expression is positively correlated with tumor stage.
| MT1G high group | MT1G low group | ||
|---|---|---|---|
| Gender | Male | 185 | 163 |
| Female | 83 | 95 | |
| Age (Median) | 62 | 59 | |
| Stage | I | 120 | 134 |
| II | 23 | 33 | |
| III | 63 | 57 | |
| IV | 51 | 32 |
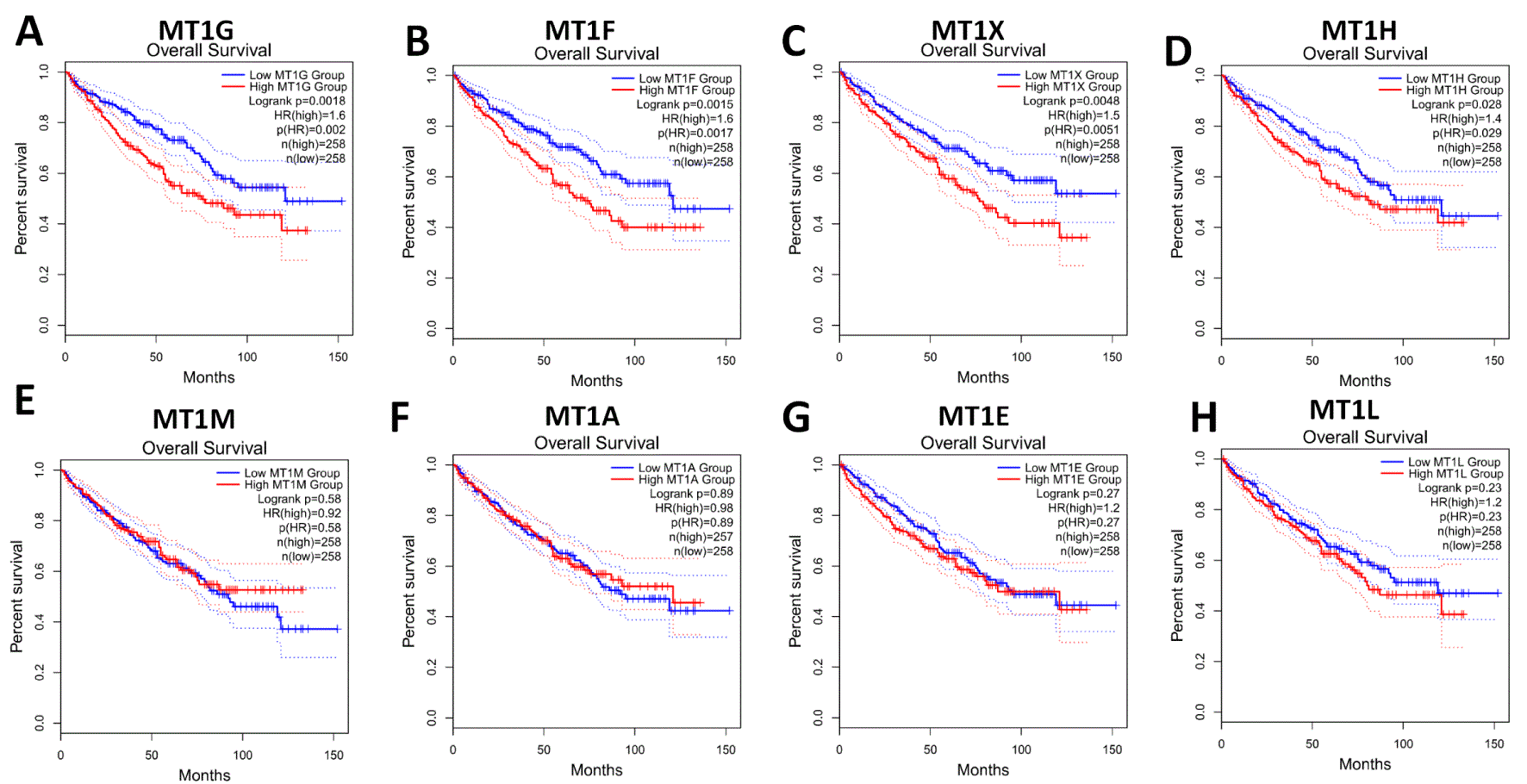
(A) MT1G, (B) MT1F, (C) MT1X, (D) MT1H, (E) MT1M, (F) MT1A, (G) MT1E and (H) MT1L. Red line represents samples with genes highly expressed and blue line for lowly expressed. HR: hazard ratio.

(A) MT1G, (B) MT1F, (C) MT1X, (D) MT1H, (E) MT1M, (F) MT1A, (G) MT1E and (H) MT1L. Red line represents samples with genes highly expressed and blue line for lowly expressed. HR: hazard ratio.
The primary goal of this study was to determine mRNA expression profile of MT1 isoforms in ccRCC tissues. Using data from TCGA-KIRC, we found that mRNA expression levels of MT1G, MT1H, MT1F, MT1X, MT1E and MT1M were simultaneously decreased in ccRCC tumor tissues compared with normal tissues. We further confirmed that MT1 isoform mRNA expression levels, especially MT1G, decreased in ccRCC tumor tissues by screening the Oncomine database. In addition, prognostic values of MT1 isoform mRNA expression in renal cancer patients were also assessed using GEPIA.
Lots of studies had demonstrated that the expression of MTs is not universal in human cancers13,14. Previous studies have shown that different MT isoform expressions were deregulated in various types of cancer11,13,27. Previous results have shown that MT isoforms are deregulated in ccRCC, hepatocellular carcinoma, prostate cancer, and papillary thyroid carcinoma6–10. In this study, we showed that MT1 isoform expression decreased in ccRCC tissues as analyzed by GEPIA based on TCGA-KIRC project and Oncomine database data, which were consistent with previous studies23,28,29. The results show that MT1 isoform expression decreased in tumor tissues, especially MT1G, which may play an important role in ccRCC development and serve as a diagnostic marker.
It’s well known that the expression of MT1 isoforms are downregulated in tumor tissues and high expression of MT1 isoforms are unfavorable prognostic characteristics and associated with advanced tumor stage13,28,29. Here, we found that MT1G, MT1F and MT1X were associated with poor prognosis in ccRCC patients. However, our data didn’t support a significant association between MT1G mRNA expression levels and renal cancer stage. In contrast, MT1F and MT1X showed positive associations between mRNA level and tumor stage, which were consistent with a previous studies that showed that MT1 isoforms are correlated with tumor stage12,18–20. The data suggested that MT1 isoforms may possibly be used as a clinical prognostic parameter.
The MT1 gene family contains MT-1A, -1B, -1E, -1F, -1G, -1H, -1M and -1X and the rest of MT-1 genes (MT-1C, -1D, -1I, -1J and -1L) are pseudogenes12,13. The expression and localization of individual MT1 isoforms and pseudogenes vary at cellular levels in individual tissues13. Based on the TCGA-KIRC transcriptome data, we couldn’t detect mRNA expression of MT1D, MT1C, MT-1I and MT1J in ccRCC, and we detected low expression of MT1B in renal cancer tissues, which meant a tissue specific expression of the MT1 isoforms. Furthermore, we confirmed that MT1G expression is strongly correlated to MT1H and MT1F and moderately correlated to MT1M, MT1X and MT1E. Considering the expression profiles of MT1 isoforms vary in different cancer tissues, further exploration of MT1 isoform expression profiles and function in ccRCC should be addressed.
In conclusion, this, to the best of our knowledge, is the first systematic study on MT1 isoform expression profiling in ccRCC and evaluation of the relationship between MT1 isoforms and clinical pathological features. However, further studies are required to understand the mechanism of downregulation of MT1 isoforms and the roles MT1 isoforms play in ccRCC, which would be of great importance to ccRCC patients.
Open Science Framework: Expression and Prognostics Significance of MT1 isoforms in Clear Cell Renal Cell Carcinoma, https://doi.org/10.17605/OSF.IO/9XUND30.
This project contains the following underlying data:
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Cancer stem cells, Cancer Biomarkers.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Cancer biology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 23 Apr 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)