Keywords
Purifying selection, diversifying selection, mismatch distribution, molecular dating, demographic history, population expansion
Purifying selection, diversifying selection, mismatch distribution, molecular dating, demographic history, population expansion
This is a minor update. Some sentences have been modified and several additional references have been included.
See the authors' detailed response to the review by Andrew G. Briscoe
See the authors' detailed response to the review by Abigail Hui En Chan and Urusa Thaenkham
Mitochondrial DNA (mtDNA) has long been a marker of choice for investigating concepts as diverse as estimating genetic diversity and effective population sizes, reconstructing species’ evolutionary histories, exploring spatial genetic subdivisions, and identifying cryptic species. All these methods assume that mtDNA variation conforms to the neutral model of molecular evolution1, but violations of this premise have long been recognised2. Over the past decades, much evidence has accumulated that mtDNA can be strongly affected by selective sweeps and background selection3–6. As a result, the usefulness of the marker in assessing genetic diversity7 and exploring spatial genetic structure in continuously distributed populations8 has been questioned, and corrections of the mitochondrial molecular clock that account for selection have been proposed9,10.
The implications of reduced genetic diversity at the species or population levels due to purifying selection has so far received little attention. When mutations in mitochondrial genes occur at fewer sites than expected under the neutral model11, molecular dating of historical demographic events by means of evolutionary rate estimates that are typically based on inter-specific divergence12,13 will result in considerable underestimates. This is particularly likely because divergence between species can be strongly affected by diversifying selection that is driven by different environmental conditions14,15, resulting in a faster accumulation of mutations characterising each species than is expected under the neutral model.
Here, we explore this issue using mitochondrial and nuclear DNA sequence data from two common southern African snails of the genus Afrolittorina that show no spatial genetic structure throughout their ranges16. The finding that data from two genetic markers with mutation rates are that assumed to differ by at least an order of magnitude17,18 have similar levels of intraspecific variation challenges the usefulness of mitochondrial DNA sequences for studying historical demographic changes.
Specimens of the snails Afrolittorina africana and A. knysnaensis were collected at 34 sites throughout South Africa (Table 1). DNA was extracted using the CTAB protocol19, amplified with universal COI primers20 and 28S primers LSU521 and LSU160022 following Williams et al.22, and sequenced on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems) using Big Dye Terminator v3.1 chemistry. Sequences were edited using MEGA723, and 28S sequences were phased in PHASE v2.1.124 using default settings. Genealogical relationships between COI haplotypes and 28S alleles were reconstructed using the median-joining algorithm25 in popArt v1.726. To explore the effect of using interspecific evolutionary rates to estimate species-level population size changes, we calculated population expansion time27 using Arlequin v3.528 using each marker’s slowest and fastest published rates for marine gastropods (Table 2).
34 sites along the South African coastline were sampled, and these are arranged from west to east.
The moment estimator τ is equal to 2ut, where u equals 2 µk (μ is the mutation rate and k is the length of the sequence), and t is the time of expansion in million of years (my).
| Species | τ | Marker | μ (%.my-1) | t (my) |
|---|---|---|---|---|
| Afrolittorina knysnaensis | 2.00 | COI | 0.501 | 0.40 (0.00 – 0.41) |
| 2.602 | 0.07 (0.00 – 0.08) | |||
| 3.25 | 28S | 0.011 | 32.1 (18.5 – 61.3) | |
| 0.052 | 6.41 (3.69 – 12.3) | |||
| Afrolittorina africana | 2.50 | COI | 0.501 | 0.50 (0.30 – 0.79) |
| 2.602 | 0.10 (0.06 – 0.15) | |||
| 2.75 | 28S | 0.011 | 27.1 (19.1 – 51.5) | |
| 0.052 | 5.42 (3.81 – 10.3) |
Species-specific genetic clusters reconstructed from COI sequences were highly distinct (Figure 1a), with a minimum number of 44 nucleotide differences between the two species’ most closely related haplotypes. In contrast, differentiation between 28S sequences (Figure 1b) was an order of magnitude smaller (4 differences).
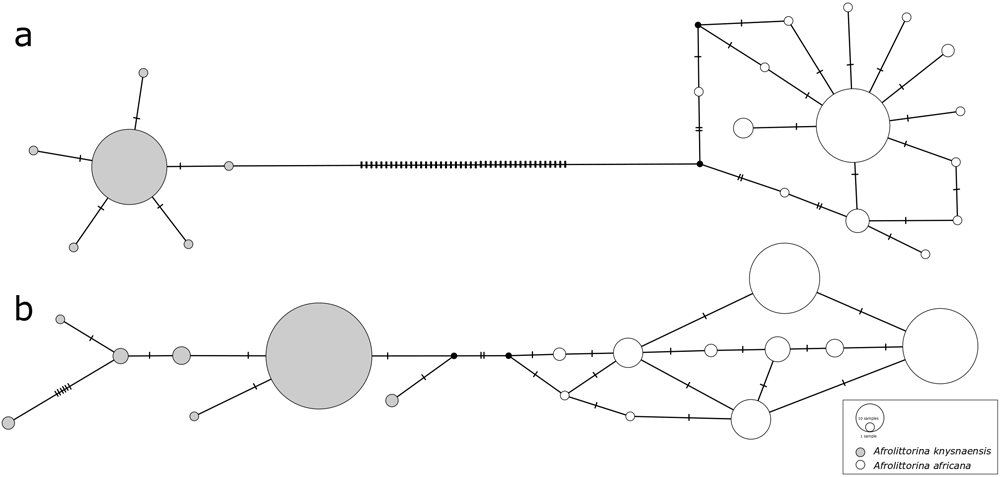
Median-joining haplotype networks constructed from a) COI sequences and b) 28S rRNA sequences of Afrolittorina knysnaensis (grey) and A. africana (white). Low intra-specific variation and high inter-specific variation of COI potentially illustrate purifying and diversifying selection, respectively. The size of circles is proportional to the frequency of each haplotype, cross-bars represent nucleotide differences, and black dots are missing haplotypes not found in the samples.
In contrast to the high inter-specific differentiation between COI haplotypes, intra-specific genetic differentiation was comparatively low for this marker, and similar to that of 28S. In A. knysnaensis, six COI haplotypes and seven 28S haplotypes were found, while the maximum differentiation between the COI haplotypes was only two nucleotide differences, but 10 for 28S. The number of haplotypes was greater for A. africana, where 14 were found for COI and 10 for 28S. Maximum nucleotide differences for this species were seven in the COI network and five for 28S.
The practical implications of two markers with very different evolutionary rates based on inter-specific divergence having similar levels of intraspecific variation are illustrated in Table 2. Using published rates, estimates of population expansion times were more than an order of magnitude greater based on the 28S data than based on the COI data.
The usefulness of the mtDNA COI gene to uncover overlooked biodiversity is undisputed because of the marker’s tendency to have a well-defined barcoding gap, as was found here. The two study species’ COI sequences were much more strongly differentiated than their 28S sequences, potentially reflecting diversifying selection as a result of adaption to different thermal environments16. In contrast, there was comparatively little genetic variation at the intraspecific level for either marker, which is likely due to the commonly reported strong purifying selection acting upon the COI gene6,9.
Many researchers explore their mtDNA sequence data for additional information, but the selective forces that together create the barcoding gap29 make its utility for other applications questionable7,8. In the present study, we have highlighted a largely unexplored problem that likely arises from selection effects in mtDNA data: the fact that demographic events using gene regions under variation-reducing purifying selection are dated using molecular clock calibrations affected by variation-increasing diversifying selection. The finding that intraspecific mtDNA variation can be as low as that of nuclear rRNA cautions against the continued use of mtDNA for exploring demographic trends by means of mismatch distributions or Bayesian skyline plots30, a practice that continues to dominate the recent literature31–34.
In our opinion, it is time to discontinue the use of fixed mtDNA rates based on divergence dating of closely related taxa, such as the closure of the Central American Seaway to date phylogenies of marine species12,13 or the 2% rule in birds35. The very large datasets generated using next-generation sequencing have considerable potential to facilitate more accurate dating by identifying nuclear markers that conform to the assumptions of the molecular clock but, curiously, fixed rates based on mtDNA data are still being used to calibrate such datasets when no suitable fossil calibration points exist36. A possible solution may involve the identification of a suite of neutral markers that can be used to assess divergence between the species used in the original molecular dating studies, and 28S rRNA may be a suitable candidate.
DNA sequences generated in this study were submitted to GenBank (COI accession numbers: MT331645–MT331814; 28S rRNA accession numbers: MT329760–MT330099).
A previous version of this article is available on bioRxiv: https://doi.org/10.1101/2020.03.31.017764.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular systematics.
Is the work clearly and accurately presented and does it cite the current literature?
No
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular systematics.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular biology, mitogenomics, phylogenetics.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 28 Aug 20 |
read | |
|
Version 1 07 May 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)