Keywords
Leymus mollis, ice-binding protein, Arctic, Alaska, Pooidae, dune grass, Chukchi Sea
Leymus mollis, ice-binding protein, Arctic, Alaska, Pooidae, dune grass, Chukchi Sea
Many revisions were made in response to reviewer comments.
Leymus mollis is not strictly an Arctic grass so the title was changed to "an Arctic population of American dunegrass, Leymus mollis".
The percent identity to L. perenne was added to the Abstract.
Details about the growing season on the Arctic coast were added to the Introduction.
We clarified the significance of facets on ice crystals in the Results and discussion section.
We added another control (Festuca) to Figure 2 to more clearly show the higher IRI activity of L. mollis.
A sentence was added to clarify how IBPs are thought to bind to ice to the Results and discussion section.
We revised the final paragraph to provide some comparative aspects of cold tolerance and adaptation in plants.
We have added a supplementary table (Table S1) as Extended data, which gives the frequency of subzero temperatures during the growing season.
See the authors' detailed response to the review by Hans Ramløv
See the authors' detailed response to the review by Matthew Carlson
See the authors' detailed response to the review by Peter L. Davies
Within the grass family (Poaceae), the subfamily Pooideae includes many cold-adapted grasses including wheat, barley and forage grasses. These grasses have developed a large family of ice-binding proteins (IBPs) that protect the plants from freezing damage (Sandve et al., 2008). The IBPs, which have negligible effect on the freezing point, are characterized by a C-terminal domain consisting of several repeating units that is associated with strong ice recrystallization inhibition (IRI) activity (Sandve et al., 2008). This region is thus called the IRI domain. Ice recrystallization, in which larger ice grains grow at the expense of smaller grains, occurs mostly at warmer sub-zero temperatures and is thought to cause damage to cell walls. Two of the most studied of these IBPs are from the grasses Lolium perenne (Sidebottom et al., 2000) and Deschampsia antarctica (John et al., 2009). Their amino acid sequences are very similar in both the N-terminal and IRI domains. The 118-a.a. IRI domain of the L. perenne IBP has been crystallized and its structure determined by X-ray crystallography (Middleton et al., 2012). The IRI domain has a beta-roll fold with eight similar coils, one side of which is the predicted ice-binding site. The spacing of the coils is very close to the repeat distance along the a-axis of ice. Interestingly, heterologous expression of the Lolium IRI domain in tomato was also shown to increase chilling (4°C) tolerance (Balamurugan et al., 2018), although the mechanism remains unclear. Here, we describe a related IBP from a pooid grass from Utqiaġvik (Barrow), Alaska, Leymus mollis. This site, at 71.3° N, is the highest latitude at which IBPs have been examined in a grass It is characterized by a short growing season from mid-June to late August with an average air temperature of 4°C (Tieszen, 1972), and frequent subzero air temperatures (Table S1, Extended data).
L. mollis, also known as American dunegrass, is found in coastal habitats, especially sand dunes across North America and Greenland. It is typically subjected to many stresses, such as low nutrient levels, salt spray, little freshwater, inundation during storms, wind abrasion, and ice storms (Gagné & Houle, 2002). Here we characterize the IBP of L. mollis and compare it with the IBP of L. perenne, a plant that is not as well adapted to such northern regions (Helgadóttir et al., 2018).
A grass sample was collected from a gravel beach at Utqiaġvik (Barrow), Alaska (71.3°N) on 6 October 2019, and stored at -80°C. The grass was shipped frozen to University of Nevada Las Vegas for analysis, but accidentally rose to ~10°C for about one day during shipment.
To obtain a sample for measuring ice structuring and IRI activities, the brown outer layers of a rhizome from just below the soil surface were peeled off, revealing green tissue underneath. About 70 mg of the green tissue was ground in 1 ml water in a mortar and pestle. The suspension was centrifuged at 14,000 rpm for 5 minutes to yield a slightly cloudy supernatant. Stem tissue from lawn grass (Festuca sp.) at the University of Nevada Las Vegas (52 mg) was similarly homogenized and used as a control. Ice-structuring activity was observed by examining the growth of an ice seed crystal with well-defined ice c- and a-axes submerged in the grass extract supernatant, as described previously (Raymond & Fritsen, 2001). Briefly, the sample was placed in a rectangular tube and the tube was submerged in a controlled temperature bath with front and rear windows. The growth of the crystal was observed at a temperature slightly below the freezing point (~-0.2°C) with a horizontally mounted dissecting microscope. Ice-structuring activity was defined as the appearance of sharply defined facets on the crystal surface. IRI activity was observed as described previously (Raymond & Fritsen, 2001). Briefly, 3 µl drops of supernatant were placed on a slide cover glass. A liquid nitrogen-cooled slide glued to a handle was pressed against the drops to form highly polycrystalline ice between the slide and cover glass. The samples were stored at -3°C in hexane and changes in recrystallization were monitored in the temperature bath described above over 21 hours. Photographs were taken through crossed polarizers.
Green tissue was obtained and homogenized as described above. DNA was extracted with a NucleoSpin Plant II kit (Machery Nagel; catalog number 740770) according to the manufacturer’s instructions. To make IBP primers, the nucleotide sequences of IBPs from L. perenne, D. antarctica and Triticum aestivum (GenBank accession numbers EU680848, FJ663044 and KU204387, respectively) were aligned. Regions with high identities at the 5’ and 3’ ends were selected for primer sequences to obtain an amplicon that covered as much of the gene as possible. Primers for 18S ribosomal RNA were selected from conserved regions in the 18S sequences from Dupontia fisheri, L. perenne and D. antarctica (GenBank accession numbers KP794861, KJ598999 and MH628292, respectively) that flanked a region of high variability. At that time, we suspected the Utqiaġvik grass sample was D. fisheri and did not use the L. mollis 18S sequence. PCR was carried out with an Eppendorf Mastercycler Personal thermal cycler, with 3 minutes initial denaturation at 95°C, followed by 35 cycles of 30 seconds denaturation at 95°C, 30 seconds annealing at 59°C and 40 seconds extension at 72°C, followed by 3 minutes final extension at 72°C using Promega GoTaq polymerase (catalog number M300B). PCR products were electrophoresed on a 2% agarose gel, stained with ethidium bromide and observed and photographed with a UVP transilluminator. IBP and 18S bands of the expected sizes were obtained (see Underlying data (Raymond, 2020c)), cleaned up with a Nucleospin gel and PCR clean-up kit (Machery Nagel, catalog number 740609) and sequenced in both directions at the UNLV Genomics Core with an Applied Biosystems 3130 sequencer. The primers that were used for sequencing are shown in Table 1.
A 3D model of the IRI domain of the Leymus IBP was predicted with SWISS-MODEL (Waterhouse et al., 2018) using the eight-coil structure of the Lolium perenne IRI domain (Protein Data Bank accession no. 3ULT) (Middleton et al., 2012) as template. SWISS MODEL proposed three structures of the Leymus IRI domain based on the template. We selected the one with the highest sequence identity (76.5%), which also was the only one that had eight orderly coils like those in the L. perenne template. The free energy of this structure was then minimized (from -42.56 to -52.58 MJ mol -1) with the YASARA Energy Minimization Server (Krieger et al., 2009) and displayed with YASARA View. Stereoviews were obtained by rotating the molecule around its vertical axis by 3°. The average distance between coils in the IRI domain was measured with the YASARA distance function as the distance between the alpha carbons of Ser15 and Ser116 divided by seven.
The obtained 18S rRNA sequence (GenBank accession no. MT506010) most closely matched the sequence of Leymus mollis (GenBank accession no. EF581964) (98.3% identity). Among the Pooidae, L. mollis is a member of the Triticodae, which includes wheat (Triticum) and barley (Hordeum), while Lolium and Deschampsia are members of the Poodae. The grass was confirmed as L. mollis from photos of the remnant spikes and leaves by Matthew Carlson (University of Alaska Anchorage) and Carolyn Parker, University of Alaska Fairbanks (personal communications to T.S.).
An extract from green rhizome tissue strongly affected the growth of an ice seed crystal, causing it to develop sharp facets (Figure 1A). The facets are an indication of the presence of ice-binding proteins. The facets themselves do not have any role in freezing tolerance, nor are they likely to form in frozen tissue; they are only a demonstration that molecules are binding to the ice surface, which is necessary for IRI. In constrast, ice grown in the presence of extract from lawn grass stem tissue (control) is smooth and rounded with no evidence of facet development (Figure 1B). The Leymus extract also had strong IRI activity. Polycrystalline ice formed from water and the lawn grass extract recrystallized significantly after 21 hours at -3°C, while the supernatant of the Leymus extract showed no recrystallization (Figure 2).

(A) Leymus mollis rhizome tissue. (B) Lawn grass (Festuca sp.) stem tissue (control). Ice seed crystals were placed in the extracts at slightly below the freezing point. Scale bars, 1 mm.
Primers based on the IBPs of other pooid grasses succeeded in amplifying a sequence that encoded the C-terminal part of the IBP gene of L. mollis, including the entire IRI domain. The sequence (GenBank accession no. MT506011) encoded 260 a.a., which corresponds to all but the first 25 a.a. of the L. perenne IBP sequence. The sequence included the stop codon and contained no introns. Although the sequence was close to the sequence of L. perenne (65% in the region of overlap), it most closely matched an IBP-like protein from Triticum aestivum (QBE94480) (86% identity, 91% similarity), in agreement with L. mollis’s classification as a member of the Triticodae. When only the IRI domains were compared, the Leymus IRI domain was also closer to the Triticum IRI domain than it was to the Lolium IRI domain. The ice-binding activities of Tritium spp. IBPs have not yet been reported.
The IRI domain of L. mollis, like that of L. perenne, has eight repeats, each consisting of 14 or 15 residues (Figure 3A). The consensus sequences of the two IBPs are virtually the same (Figure 3A).
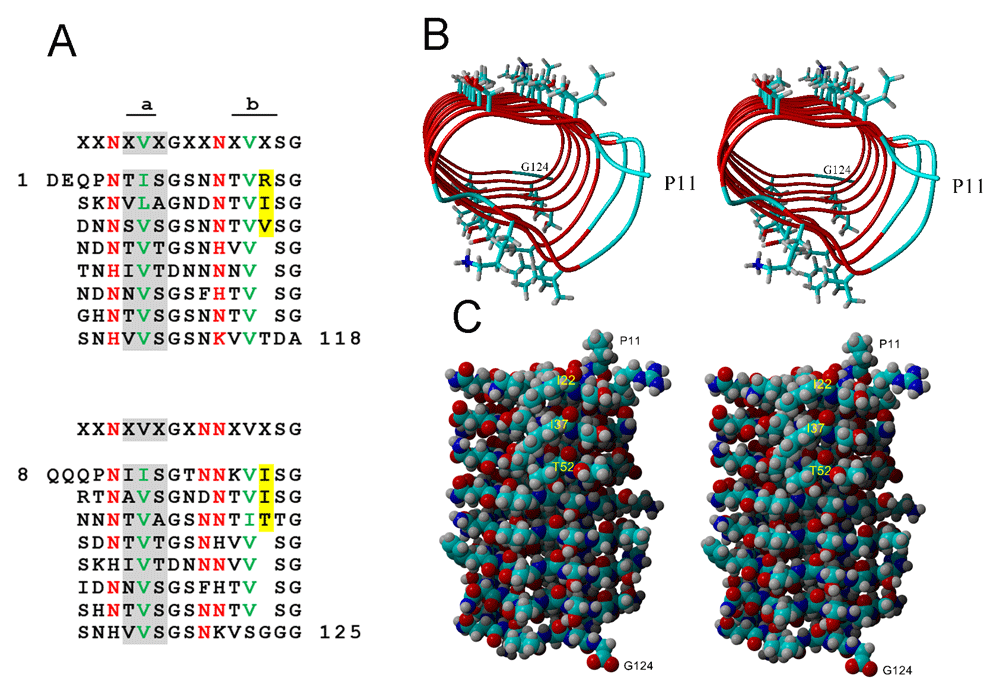
(A) Comparison of the Leymus repeats (bottom) with those in the IRI domain of Lolium perenne (top). The Lolium data and the color scheme have been reproduced with permission from Middleton et al. (2012). Both domains have eight repeats. Consensus sequences are shown on top. Numbers indicate a.a. residue numbers. In the Lolium sequence, the gray background indicates the ice-binding site called the a side and the yellow background indicates a bulge on the b side. Similar features are found in the Leymus sequence. (B, C) Stereoviews predicted by SWISS MODEL using the IRI domain of L. perenne as template. SWISS MODEL was able to model the Leymus structure from Pro11 to Gly124, which corresponds to Asp1 to Ala118 in L. perenne. (B) View through the center of the coils, in which the ice-binding site (the a side) is on top. Amino acid side chains are shown only for the a and b sides. Color code of the ribbon: red, beta strand; cyan, coil. Color code of amino acid side chain: Cyan, C; blue, N, red, O; gray, H. (C) View of the b side of a space-filling model. A bulge is created by two residues on each of the first three coils. The first of the two residues in each coil is labeled. Colors are the same as those for the side chains in A.
The predicted structure of the L. mollis IBP IRI domain, obtained from using the L. perenne IBP IRI domain structure as template, resembles the Lolium IRI domain in several respects: it has eight repeats corresponding to eight coils in a beta-roll fold (Figure 3B); a flat side (the a side) that is populated with two rows that are rich in Thr and Ser residues (Figure 3B); an irregular side (the b side) that has a bulge in the first three coils (Figure 3C); and an interior that is dominated by Asn/His ladders (red residues in Figure 3A). The a side has been identified as the ice-binding site of L. perenne IBP (Middleton et al., 2012). In Lolium, the average spacing between the alpha carbons of the row of amino acids on the ice-binding site is 4.5 Å, which is very similar to the spacing along the a-axis of ice (4.51 Å). In Leymus, the average spacing was calculated as 4.77 Å. The regular array of hydrophilic residues is thought to order water molecules that act as a glue between the IBP and the ice (Middleton et al., 2012).
In summary, we describe an ice-binding protein from the grass Leymus mollis with ice-structuring and ice recrystallization inhibition (IRI) activities. The collection site of the grass, a gravelly beach on the Chukchi Sea, is an extreme habitat subject to numerous environmental stresses. L. mollis appears to be better adapted to high latitudes than L. perenne, which does not grow well north of 60°N (Helgadóttir et al., 2018). It seems unlikely that the greater cold hardiness of Leymus can be attributed to its IBP as the IBPs of the two grasses are very similar. However, many proteins contribute to the cold hardiness of forage grasses, including transcription factors, cell membrane proteins, and proteins involved in cell signalling, cellular transport and photosynthesis (Sandve et al., 2011). It is thus likely that many of these factors are responsible for L. mollis’s survival in the high Arctic. One approach to identifying them would be a comparison of transcriptomes of the Arctic population with a California population that never experiences freezing conditions.
Leymus mollis 18S ribosomal sequence on GenBank, Accession number MT506010
Leymus mollis IBP sequence on GenBank, Accession number MT506011
Figshare: Ice structuring by ice-binding protein of Leymus mollis DSCN5945.JPG. https://doi.org/10.6084/m9.figshare.12401966.v1 (Raymond, 2020a)
Figshare: Leymus mollis ice recrsytallization inhibition. https://doi.org/10.6084/m9.figshare.12401954.v1 (Raymond, 2020b)
Figshare: PCR amplification of IBP and 18S genes of Leymus mollis. https://doi.org/10.6084/m9.figshare.12401957.v1 (Raymond, 2020c)
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Figshare: Table S1. Number of days with subzero air temperatures during the growing season at Utqiaġvik, AK. https://doi.org/10.6084/m9.figshare.12765983.v1 (Raymond & Sformo, 2020)
This project contains the following extended data:
Table S1.docx - Number of days with subzero air temperatures during the growing season at Utqiaġvik, AK. Data provided by the National Climatic Data Center.
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
JR thanks the School of Life Sciences, University of Nevada Las Vegas, for providing facilities for carrying out this study.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Cold tolerance of ectothermic animals, antifreeze proteins, cryptobiosis
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Plant ecology and evolution - emphasis on Arctic and Boreal systems
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Protein biochemistry, structural biology, ice-binding proteins
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 17 Aug 20 |
|||
|
Version 1 26 Jun 20 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)