Introduction
Exonic circular RNAs (circRNAs) constitute a large class of regulatory non-coding endogenous RNAs with variable composition. Over the past few years, research into their biogenesis and biological function has exploded. First discovered in viroids, where they appear as circular genomes1, circRNAs have been shown to exist in all kingdoms of life, with thousands of circRNAs identified across species from archaea to humans2,3. For decades, circRNAs were considered to be extremely rare in nature and, in particular in eukaryotes, they were seen as minor RNA structural variants attributed to transcriptional noise4. Owing to progress in analytical techniques and the development of specific methodologies for the discovery and identification of circRNAs (recently reviewed in 5), this picture has dramatically changed over the past several years. It became obvious that circRNAs are abundant, evolutionarily conserved, and stable species in all eukaryotes studied today, although some eukaryotes like Saccharomyces cerevisiae have only very few circRNAs because of their few multi-intronic genes. The biogenesis and full functional repertoire of circRNAs have not yet been fully elucidated. Here we will review recent progress in circRNA research, focussing on new data regarding their biogenesis, cellular function, and involvement in diseases. We will extend our view to strategies for controlled generation of circRNAs in vivo and in vitro and discuss putative applications. We do not include the development of tools and biochemical methods for the accurate identification and characterization of circRNAs, since this, as mentioned above, has been extensively reviewed very recently5.
Biogenesis of circRNAs
Most circRNAs are expressed from known protein-coding genes and are composed of single or multiple exons3. They are produced by backsplicing, a process that occurs in a reversed orientation as compared with canonical splicing. Hence, instead of joining an upstream 5'-splice site with a downstream 3'-splice site in a sequential order to produce a linear RNA, a downstream 5'-splice site is linked to an upstream 3'-splice site to yield a circRNA (Figure 1a)6–9. Still, the formation of circRNA was shown to be dependent on the canonical splicing machinery, making backsplicing a process that competes with canonical splicing10,11. In addition to exonic circRNAs, circular RNAs containing sequences from introns (ciRNA) and circRNAs containing sequences from exons with introns retained between the exons (exon-intron circRNA or short IEciRNA) have been found. ciRNAs presumably result from intron lariats that escaped de-branching during canonical splicing and do not belong to circRNAs. They reside in the nucleus, where they may control the transcription of their parental genes12,13. However, previous work also suggests that some ciRNAs are stable in the cytoplasm14,15. Exonic circRNAs localize to the cytoplasm, where they are exported from the nucleus in a length-dependent manner16. In general, all exons found in linear transcripts may appear in circRNAs. However, it is also possible that circRNAs contain exons which do not appear in linearly spliced transcripts17.
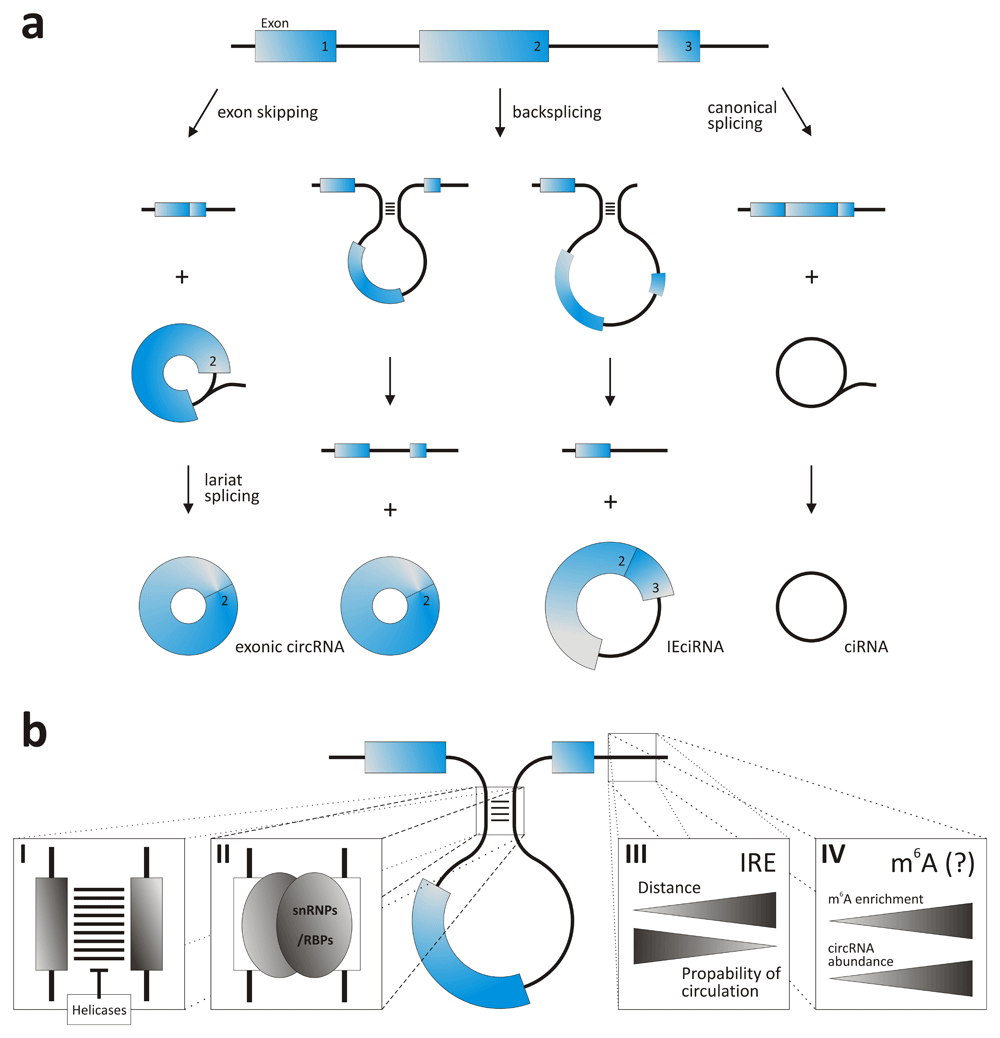
Figure 1. Biogenesis of circRNAs.
(a) Modes of circRNA formation. (b) Factors supporting backsplicing: inverted repeat sequences (I), binding sites for RBPs of RNPs (II), IRE distance (III), and m6A-enriched sites (IV). For further explanation, see main text. ciRNA, circular RNA containing sequences from introns; circRNA, circular RNA; IEciRNA, circRNA containing sequences from exons with introns retained between the exons; IRE, inverted repeat; RBP, RNA-binding protein; snRNP, small nuclear ribonucleoprotein.
Successful backsplicing requires the splice sites to be brought into proximity (Figure 1b). This often is supported by inverted repeats (IRE), especially Alu elements, flanking the exons to be circularized and allowing multiple circRNAs to be produced from a single gene8,18,19. In addition to IRE, interaction of the precursor mRNA with ribonucleoproteins (RNPs) or proteins was found to support circRNA formation11,20–23. Other RNA-binding proteins (RBPs) that support circularization are, for example, the heterogeneous nuclear RNP L (HNRNPL)24, double-stranded RNA-binding domain containing immune factors NF90/NF11025, or DHX9, an abundant nuclear RNA helicase26. Moreover, circRNA biogenesis underlies the combinatorial control of splice factors21 and can also be suppressed by helicases6,27. Pre-mRNA structure plays an important role, as flanking sequences (e.g. IRE or RNP-binding sites) or the distance between splice sites is most important28. Furthermore, N6-methylation of adenosine can promote circRNA biogenesis, as it was recently shown that m6A-enriched sites guide backsplicing in male germ cells29. CircRNA levels are also modulated by the levels of core spliceosome components30, and it was suggested that the same spliceosome can assemble across an exon and that it either remodels to span an intron for canonical linear splicing or catalyses backsplicing to generate circRNA31.
Backsplicing is less efficient than linear splicing32, and, typically, circRNAs are produced at a lower level than their linear counterparts. Yet circRNAs may be the more abundant isoform in specific cells and tissues33,34, which may be attributed to their higher stability. Owing to the covalently closed ring structure, circRNAs are resistant to degradation by exonucleases, thus undergoing slower turnover. The higher stability implies that possible functions of circRNA may be associated with their longer lifespan. Nevertheless, there is evidence of circRNA turnover, as it was shown that upon poly(I:C) stimulation or viral infection, circRNAs are globally degraded by RNase L, a process required for PKR activation in early cellular innate immune responses35. Furthermore, m6A-containing circRNAs, when bound to the m6A reader protein YTHDF2, become rapidly degraded by the RNase P/MRP complex36. circRNA degradation is also mediated via a structure-related RNA decay pathway that is independent of specific single-stranded sequences but recognizes double-stranded structures in the 3' UTR of mRNAs, as well as highly structured circRNAs37.
Biological functions of circRNAs
To date, biological function has been investigated for only a minor fraction of circRNAs. Many of those have been proposed to act as miRNA sponges38–42 or protein sponges11,43. In addition, circRNAs may enhance protein function13,33, assist protein target interaction44–46, or recruit proteins to specific locations47. An early example for a potential miRNA sponge is circRNA ciRS-7, also known as CDR1as, comprising over 70 binding sites for miR-740. However, this function is still controversially discussed, in particular when looking at stoichiometric ratios of the target sequences to the number of binding sites in the circRNA48,49. Furthermore, analysis of 7,000 human circRNAs revealed that most of them are not enriched in miRNA-binding sites3.
Some circRNAs possess binding sites for specific proteins, which upon binding lose interaction with other targets50. In a similar manner, circRNAs have been described to function as protein scaffolds, assisting the assembly of protein complexes43,44,46,51. For example, circFoxo3 was shown to inhibit the progression of the cell cycle by formation of a ternary complex with CDK2 and p21, thereby acting as a tumor suppressor46, or to specifically recruit the ubiquitinylation system, thus triggering degradation of mutated p53 by the proteasome complex44. CircRNA can also regulate the subcellular localization of specific proteins, as shown for circ-Amotl1 binding to Stat3, AKT1, and PDK145,52. Because backsplicing competes with canonical splicing, the formation of circRNAs is also considered to be a mode of regulating the expression of a specific gene. The protein Muscleblind (MBL) binds to the flanking introns of circMBL derived from the muscleblind gene by backsplicing. As a result, MBL levels are modulated, which in turn strongly affects circMBL biosynthesis43.
There has been some indication that cells can differentiate between endogenous and exogenous circRNA. Exogenously introduced circRNA was shown to have a stimulating effect on the immune system because it is recognized by the pattern recognition receptor retinoic acid inducible gene I (RIG-I), thereby eliciting a strong immune response. Apparently, this applies only to unmodified circRNA because m6A-modified circRNA was shown to inhibit innate immunity53. Endogenous circRNA, on the contrary, did not show such an effect. Based on this observation, it was proposed that endogenous circRNA is recognized as self, owing to the identity of its flanking introns that led to circularization54. More recent findings, however, are contradictory, as they suggest that unmodified exogenous circRNA is able to bypass cellular RNA sensors and thus does not induce an immune response in RIG-I and Toll-like receptor (TLR) competent cells and in mice55. Endogenous circRNAs can collectively bind and suppress activation of the double-stranded RNA (dsRNA)-activated protein kinase PKR, thereby controlling innate immune responses56. As already mentioned above, double-stranded RNA-binding domain-containing immune factors NF90/NF110 are key regulators in circRNA biogenesis, pointing to the role of circRNAs in immunity. Upon viral infection, circRNA expression is decreased, and NF90/NF110 released from circRNP complexes bind to viral mRNAs as part of their functions in antiviral immune response25.
Interestingly, some circRNAs containing internal ribosome entry site (IRES) elements and AUG sites may be translated into unique peptides under specific conditions, in particular upon cellular stress52,57–61, although the functional relevance of the majority of circRNA-derived peptides is not yet known. Earlier studies had suggested that circRNAs might be translated without the existence of an IRES sequence, following the so-called rolling circle translation mechanism62. In more recent studies, however, translation of circRNAs was shown to be dependent on the presence of different IRESs (either viral IRES sequences36,37 or m6A28). Yet it should be noted that even though several studies have reported cases of circRNA translation, others have completely failed to find evidence63.
Taken together, circRNAs appear to play a regulatory role in different levels of gene expression, which also explains their association with diverse diseases, pathological conditions, and expression patterns specific for certain cell types and tissues.
CircRNAs in diseases
CircRNAs have been associated with the initiation and progression of several diseases, including cancer, neurodegenerative diseases, cardiovascular diseases, and diabetes51,64–69, and thus have also been considered as biomarkers for disease prognostics and diagnostics and as targets or tools for disease treatment70–72. There is also indication of circRNAs accumulating with aging73–75. Work in the field is currently centered around screening for and identifying disease-associated circRNAs, whereas the underlying mechanisms of action remain mostly unknown. In particular, the involvement of circRNAs in cancer development and progression is obvious, as numerous circRNAs have been discovered to upregulate or downregulate gene expression in cancer tissues and promote cancer cell reproduction35,66,76–83. Over the past two years, numerous circRNAs have been shown to affect cell proliferation, invasion, migration, and apoptosis and have been suggested to act as therapeutic targets or biomarkers for diagnosis and prognosis in various types of cancers84–90. There have been indications of circRNAs occurring in the tumor microenvironment91 and in exosomes92, with their role in cell-to-cell communication and spreading of pathological processes continuing to be unveiled92–94. Recent results have shown that circRNA-loaded exosomes promote cell proliferation and invasion in colorectal95 and prostate96 cancer. It has been suggested that the effect of extracellular circRNAs can be reversed by the addition of siRNAs targeting those circRNAs, hence making it a promising therapeutic strategy96,97.
Standing out in the majority of research of "more classical" diseases is the increasing knowledge of the roles of circRNAs in aging, where age-related changes in splicing, and thus in the level of lncRNAs and circRNAs, are discussed98. Furthermore, it has been found that the expression of circRNAs can be sensitive to different types of pollution (organic, heavy metal, and others) and therefore might be used as a biomarker or prevention/treatment target for pollution-induced diseases99.
Strategies for controlled generation of circRNAs in vitro and in vivo
Several methods for controlled generation of circRNAs based on chemical or enzymatic/ribozymatic strategies have been investigated100,101. Circularization can be performed either in vivo or in vitro. For direct production of circRNA in cell culture, usually the sequence of interest is cloned into an artificial exon that is flanked by complementary intronic repeats. The plasmid is then transfected into cells, transcription is induced, and the cellular splicing machinery generates the desired circRNA19,102 (Figure 2a). Alternatively, the sequence of interest can be cloned in between a permuted self-splicing intron (permuted intron exon [PIE] strategy, see below), such that circularization occurs by the inherent ribozyme activity of the intron103,104. An expression vector comprising such self-splicing introns is also suited for circularization in vitro by linearization of the plasmid and subsequent in vitro transcription of the linearized template. The formed transcript undergoes circularization by its self-splicing activity103.

Figure 2. Strategies for circRNA generation in vitro and in vivo.
(a) Overexpression vector, (b) chemical or enzymatic ligation, (c) circularization via Tornado system, (d) permuted intron exon (PIE) strategy, (e) induction of backsplicing by Csy4, and (f) generation of circRNA using tRNA splicing mechanism. For further explanation, see main text. BHB, bulge-helix-bulge; IRES, internal ribosome entry site; TSEN, tRNA splicing endonuclease complex.
Chemical ligation methods can be applied only for in vitro circularization. Linear RNA obtained from chemical synthesis or in vitro transcription and phosphorylated at the 3'- or 5'- terminus can be intramolecularly ligated with the help of condensing agents100. In addition, enzymatic ligation with DNA or RNA ligases is an option (Figure 2b)100,101.
Recently, a seminal approach for circRNA production in vivo, called Tornado (twister optimized RNA for durable overexpression), was introduced (Figure 2c)105. The twister ribozyme is employed in a combined approach with the cellular RtcB ligase. The ribozymes flanking the sequence to be circularized generate by cutting themselves off the 5'-terminal OH and 3'-terminal phosphate required by the cellular RtcB ligase to perform the following ligation step.
Already known for a while and newly moved into focus by recent studies is the PIE strategy, which uses a group I self-splicing intron (also a ribozyme) for the production of a circRNA either in vitro or in vivo106. The two intron halves (5'- and 3'- intron) flanking the exon are arranged in a permuted manner, such that during splicing a circularized exon and two linear intron halves are formed (Figure 2d). The PIE strategy was successfully applied for RNA sequences up to five kilobases, and a PIE-produced circRNA carrying an IRES sequence was shown to be successfully translated in cells52,104,107.
Another possibility to selectively circularize RNA sequences is utilizing the tRNA splicing machinery108,109. A tRNA precursor is specifically recognized by the tRNA splicing endonuclease complex (TSEN) based on a bulge-helix-bulge (BHB) motif, then cleaved and ligated by a ligase, yielding the mature tRNA and a circularized intron (Figure 2f). A desired sequence can be introduced in such a construct between the two intron halves to become circularized upon tRNA splicing. Still another method exploiting the cell’s own splicing machinery for circularization is the system based on RNA cleavage by the CRISPR endonuclease Csy4110. Csy4 recognizes a 16-nucleotide hairpin in RNA and specifically cleaves off the RNA downstream of that hairpin region. The protein is utilized for RNA circularization to cleave a site in a defined intron, thereby removing a competing downstream splice site, which otherwise would interfere with backsplicing, and thus inducing formation of the desired circRNA (Figure 2e)110.
Application of circRNAs
After research in the field of circRNA was dominated by their identification and studies into biogenesis and function, reports on the application of circRNAs have started to emerge more recently. Because of their stability and association with diseases, endogenous circRNAs are potential candidates as biomarkers or therapeutic targets111–113. Likewise, exogenous circRNAs can be introduced into cells to fulfil a defined function. Several feasible concepts for the therapeutic application of circRNAs have already been discussed and to some extent successfully implemented. An obvious possibility for the application of circRNAs is the development of designed miRNA sponges. An artificial circRNA molecule comprising multiple binding sites for miRNA-122, which plays an essential role in the life cycle of the hepatitis C virus, was successfully used to inhibit the synthesis of viral proteins in the host cell114. In a similar way, the activity of specific proteins in the cellular context was controlled by circularized aptamers105. Moreover, circular aptamers have shown great potential as intracellularly expressed biosensors for defined metabolites103,105.
Because some circRNAs play a role in alternative splicing and transcription, it is feasible to use them for the regulation of those processes within the cell, thereby driving gene expression in the direction of specific transcription and splicing products. In addition, circularization of RNA opens up the opportunity to apply RNA therapeutics that are administered as a linear construct until now (for example, mRNA vaccines) in a circular form, thereby significantly increasing their stability. If the circRNA additionally possesses an IRES sequence, translation of that RNA is possible, whereby therapeutic proteins may be expressed directly in target cells. Because of results suggesting that circRNAs can activate the immune system via the RIG-I pathway, it is also feasible to employ exogenous circRNA as an adjuvant in vaccines to elicit a more efficient immune response upon vaccination54.
In all of the approaches described above, it has to be taken into consideration that side effects may arise as a result of the applied circRNA. For example, expression of the desired circRNA from an overexpression vector or translation of a protein encoded by the circRNA can significantly vary dependent on the respective cell type115. In some cases, the formation of linear RNA concatemers by rolling circle transcription was also observed112. Those concatemers can lead to toxic effects within the cell. The function of immune activator mentioned above can also be a drawback of circRNA if the RNA is to be applied in a context wherein an immune response is not desired.
Conclusions
The occurrence of circRNAs in all kingdoms of life has been demonstrated, and it is beyond doubt that these abundant stable RNA species play important biological roles. The elucidation of circRNA function has included the development of methods for circRNA identification and characterization and of strategies for circRNA generation. It has become clear that circRNAs are strongly involved in diseases, although their action is enormously multifaceted. Even with all of the effort over the past decade to shed light onto this still-emerging field, the intracellular and extracellular roles of circRNAs as well as their functional role in bigger networks with other RNAs and proteins require ongoing endeavor to gain full understanding, and with that the opportunity to use circRNAs as biomarkers or therapeutic agents and targets.
Abbreviations
ciRNA, circular RNA containing sequences from introns; circRNA, circular RNA; IRE, inverted repeats; IRES, internal ribosome entry site; MBL, muscleblind; PIE, permuted intron exon; RIG-I, retinoic acid inducible gene I; RNP, ribonucleoprotein.
Faculty Opinions recommendedReferences
- 1.
Flores R, Grubb D, Elleuch A, et al.:
Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: Variations on a theme.
RNA Biol.
2011; 8(2): 200–6. PubMed Abstract
| Publisher Full Text
- 2.
Danan M, Schwartz S, Edelheit S, et al.:
Transcriptome-wide discovery of circular RNAs in Archaea.
Nucleic Acids Res.
2012; 40(7): 3131–42. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 3.
Guo JU, Agarwal V, Guo H, et al.:
Expanded identification and characterization of mammalian circular RNAs.
Genome Biol.
2014; 15(7): 409. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 4.
Cocquerelle C, Mascrez B, Hétuin D, et al.:
Mis-splicing yields circular RNA molecules.
FASEB J.
1993; 7(1): 155–60. PubMed Abstract
| Publisher Full Text
- 5.
Kristensen LS, Andersen MS, Stagsted LVW, et al.:
The biogenesis, biology and characterization of circular RNAs.
Nat Rev Genet.
2019; 20(11): 675–91. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 6.
Ivanov A, Memczak S, Wyler E, et al.:
Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals.
Cell Rep.
2015; 10(2): 170–7. PubMed Abstract
| Publisher Full Text
- 7.
Kelly S, Greenman C, Cook PR, et al.:
Exon Skipping Is Correlated with Exon Circularization.
J Mol Biol.
2015; 427(15): 2414–7. PubMed Abstract
| Publisher Full Text
- 8.
Zhang XO, Wang HB, Zhang Y, et al.:
Complementary sequence-mediated exon circularization.
Cell.
2014; 159(1): 134–47. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 9.
Barrett SP, Wang PL, Salzman J:
Circular RNA biogenesis can proceed through an exon-containing lariat precursor.
eLife.
2015; 4: e07540. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 10.
Starke S, Jost I, Rossbach O, et al.:
Exon circularization requires canonical splice signals.
Cell Rep.
2015; 10(1): 103–11. PubMed Abstract
| Publisher Full Text
- 11.
Ashwal-Fluss R, Meyer M, Pamudurti NR, et al.:
circRNA Biogenesis Competes with Pre-mRNA Splicing.
Mol Cell.
2014; 56(1): 55–66. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 12.
Li Z, Huang C, Bao C, et al.:
Exon-intron circular RNAs regulate transcription in the nucleus.
Nat Struct Mol Biol.
2015; 22(3): 256–64. PubMed Abstract
| Publisher Full Text
- 13.
Zhang Y, Zhang XO, Chen T, et al.:
Circular Intronic Long Noncoding RNAs.
Mol Cell.
2013; 51(6): 792–806. PubMed Abstract
| Publisher Full Text
- 14.
Talhouarne GJS, Gall JG:
Lariat intronic RNAs in the cytoplasm of vertebrate cells.
Proc Natl Acad Sci U S A.
2018; 115(34): E7970–E7977. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 15.
Talhouarne GJS, Gall JG:
Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes.
RNA.
2014; 20(9): 1476–87. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 16.
Huang C, Liang D, Tatomer DC, et al.:
A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs.
Genes Dev.
2018; 32(9–10): 639–44. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 17.
Zhang XO, Dong R, Zhang Y, et al.:
Diverse alternative back-splicing and alternative splicing landscape of circular RNAs.
Genome Res.
2016; 26(9): 1277–87. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 18.
Jeck WR, Sorrentino JA, Wang K, et al.:
Circular RNAs are abundant, conserved, and associated with ALU repeats.
RNA.
2013; 19(2): 141–57. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 19.
Liang D, Wilusz JE:
Short intronic repeat sequences facilitate circular RNA production.
Genes Dev.
2014; 28(20): 2233–47. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 20.
Conn SJ, Pillman KA, Toubia J, et al.:
The RNA binding protein quaking regulates formation of circRNAs.
Cell.
2015; 160(6): 1125–34. PubMed Abstract
| Publisher Full Text
- 21.
Kramer MC, Liang D, Tatomer DC, et al.:
Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins.
Genes Dev.
2015; 29(20): 2168–82. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 22.
Nasim FUH, Hutchison S, Cordeau M, et al.:
High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5' splice site selection in support of a common looping out and repression mechanism.
RNA.
2002; 8(8): 1078–89. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 23.
Lamichhane R, Daubner GM, Thomas-Crusells J, et al.:
RNA looping by PTB: Evidence using FRET and NMR spectroscopy for a role in splicing repression.
Proc Natl Acad Sci U S A.
2010; 107(9): 4105–10. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 24.
Fei T, Chen Y, Xiao T, et al.:
Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing.
Proc Natl Acad Sci U S A.
2017; 114(26): E5207–E5215. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 25.
Li X, Liu CX, Xue W, et al.:
Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection.
Mol Cell.
2017; 67(2): 214–227.e7. PubMed Abstract
| Publisher Full Text
- 26.
Aktaş T, Avşar Ilık İ, Maticzka D, et al.:
DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome.
Nature.
2017; 544(7648): 115–9. PubMed Abstract
| Publisher Full Text
- 27.
Ottesen EW, Luo D, Seo J, et al.:
Human Survival Motor Neuron genes generate a vast repertoire of circular RNAs.
Nucleic Acids Res.
2019; 47(6): 2884–905. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 28.
Welden JR, Stamm S:
Pre-mRNA structures forming circular RNAs.
Biochim Biophys Acta Gene Regul Mech.
2019; 1862(11–12): 194410. PubMed Abstract
| Publisher Full Text
- 29.
Tang C, Xie Y, Yu T, et al.:
m6A-dependent biogenesis of circular RNAs in male germ cells.
Cell Res.
2020; 30(3): 211–28. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 30.
Liang D, Tatomer DC, Luo Z, et al.:
The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting.
Mol Cell.
2017; 68(5): 940–954.e3. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 31.
Li X, Liu S, Zhang L, et al.:
A unified mechanism for intron and exon definition and back-splicing.
Nature.
2019; 573(7774): 375–80. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 32.
Zhang Y, Xue W, Li X, et al.:
The Biogenesis of Nascent Circular RNAs.
Cell Rep.
2016; 15(3): 611–24. PubMed Abstract
| Publisher Full Text
- 33.
Salzman J, Gawad C, Wang PL, et al.:
Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types.
PLoS One.
2012; 7(2): e30733. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 34.
Izuogu OG, Alhasan AA, Mellough C, et al.:
Analysis of human ES cell differentiation establishes that the dominant isoforms of the lncRNAs RMST and FIRRE are circular.
BMC Genomics.
2018; 19(1): 276. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 35.
Liu J, Li D, Luo H, et al.:
Circular RNAs: The star molecules in cancer.
Mol Aspects Med.
2019; 70: 141–52. PubMed Abstract
| Publisher Full Text
- 36.
Park OH, Ha H, Lee Y, et al.:
Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex.
Mol Cell.
2019; 74(3): 494–507.e8. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 37.
Fischer JW, Busa VF, Shao Y, et al.:
Structure-Mediated RNA Decay by UPF1 and G3BP1.
Mol Cell.
2020; 78(1): 70–84.e6. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 38.
Piwecka M, Glažar P, Hernandez-Miranda LR, et al.:
Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function.
Science.
2017; 357(6357): eaam8526. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 39.
Zheng Q, Bao C, Guo W, et al.:
Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs.
Nat Commun.
2016; 7: 11215. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 40.
Hansen TB, Jensen TI, Clausen BH, et al.:
Natural RNA circles function as efficient microRNA sponges.
Nature.
2013; 495(7441): 384–8. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 41.
Memczak S, Jens M, Elefsinioti A, et al.:
Circular RNAs are a large class of animal RNAs with regulatory potency.
Nature.
2013; 495(7441): 333–8. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 42.
Hausser J, Zavolan M:
Identification and consequences of miRNA-target interactions--beyond repression of gene expression.
Nat Rev Genet.
2014; 15(9): 599–612. PubMed Abstract
| Publisher Full Text
- 43.
Abdelmohsen K, Panda AC, Munk R, et al.:
Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1.
RNA Biol.
2017; 14(3): 361–9. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 44.
Du WW, Fang L, Yang W, et al.:
Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity.
Cell Death Differ.
2017; 24(2): 357–70. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 45.
Zeng Y, Du WW, Wu Y, et al.:
A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair.
Theranostics.
2017; 7(16): 3842–55. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 46.
Du WW, Yang W, Liu E, et al.:
Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2.
Nucleic Acids Res.
2016; 44(6): 2846–58. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 47.
Chen N, Zhao G, Yan X, et al.:
A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1.
Genome Biol.
2018; 19(1): 218. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 48.
Denzler R, Agarwal V, Stefano J, et al.:
Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance.
Mol Cell.
2014; 54(5): 766–76. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 49.
Thomson DW, Dinger ME:
Endogenous microRNA sponges: Evidence and controversy.
Nat Rev Genet.
2016; 17(5): 272–83. PubMed Abstract
| Publisher Full Text
- 50.
Luo J, Liu H, Luan S, et al.:
Guidance of circular RNAs to proteins’ behavior as binding partners.
Cell Mol Life Sci.
2019; 76(21): 4233–43. PubMed Abstract
| Publisher Full Text
- 51.
Holdt LM, Stahringer A, Sass K, et al.:
Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans.
Nat Commun.
2016; 7: 12429. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 52.
Yang Y, Fan X, Mao M, et al.:
Extensive translation of circular RNAs driven by N6-methyladenosine.
Cell Res.
2017; 27(5): 626–41. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 53.
Chen YG, Chen R, Ahmad S, et al.:
N6-Methyladenosine Modification Controls Circular RNA Immunity.
Mol Cell.
2019; 76(1): 96–109.e9. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 54.
Chen YG, Kim MV, Chen X, et al.:
Sensing Self and Foreign Circular RNAs by Intron Identity.
Mol Cell.
2017; 67(2): 228–238.e5. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 55.
Wesselhoeft RA, Kowalski PS, Parker-Hale FC, et al.:
RNA Circularization Diminishes Immunogenicity and Can Extend Translation Duration In Vivo.
Mol Cell.
2019; 74(3): 508–520.e4. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 56.
Liu CX, Li X, Nan F, et al.:
Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity.
Cell.
2019; 177(4): 865–880.e21. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 57.
Zhang M, Zhao K, Xu X, et al.:
A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma.
Nat Commun.
2018; 9(1): 4475. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 58.
Zhang M, Huang N, Yang X, et al.:
A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis.
Oncogene.
2018; 37(13): 1805–14. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 59.
Pamudurti NR, Bartok O, Jens M, et al.:
Translation of CircRNAs.
Mol Cell.
2017; 66(1): 9–21.e7. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 60.
Legnini I, Di Timoteo G, Rossi F, et al.:
Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis.
Mol Cell.
2017; 66(1): 22–37.e9. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 61.
Begum S, Yiu A, Stebbing J, et al.:
Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas.
Oncogene.
2018; 37(30): 4055–7. PubMed Abstract
| Publisher Full Text
- 62.
Abe N, Matsumoto K, Nishihara M, et al.:
Rolling Circle Translation of Circular RNA in Living Human Cells.
Sci Rep.
2015; 5: 16435. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 63.
Stagsted LVW, Nielsen KM, Daugaard I, et al.:
Noncoding AUG circRNAs constitute an abundant and conserved subclass of circles.
Life Sci Alliance.
2019; 2(3): e201900398. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 64.
Fang Y, Wang X, Li W, et al.:
Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus.
Int J Mol Med.
2018; 42(4): 1865–74. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 65.
Hanan M, Soreq H, Kadener S:
CircRNAs in the brain.
RNA Biol.
2017; 14(8): 1028–34. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 66.
Kristensen LS, Hansen TB, Venø MT, et al.:
Circular RNAs in cancer: Opportunities and challenges in the field.
Oncogene.
2018; 37(5): 555–65. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 67.
Li H, Li K, Lai W, et al.:
Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus.
Clinica Chimica Acta.
2018; 480: 17–25. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 68.
Aufiero S, Reckman YJ, Pinto YM, et al.:
Circular RNAs open a new chapter in cardiovascular biology.
Nat Rev Cardiol.
2019; 16(8): 503–14. PubMed Abstract
| Publisher Full Text
- 69.
Vo JN, Cieslik M, Zhang Y, et al.:
The Landscape of Circular RNA in Cancer.
Cell.
2019; 176(4): 869–881.e13. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 70.
Li P, Chen S, Chen H, et al.:
Using circular RNA as a novel type of biomarker in the screening of gastric cancer.
Clin Chim Acta.
2015; 444: 132–6. PubMed Abstract
| Publisher Full Text
- 71.
Bahn JH, Zhang Q, Li F, et al.:
The Landscape of MicroRNA, Piwi-Interacting RNA, and Circular RNA in Human Saliva.
Clin Chem.
2015; 61(1): 221–30. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 72.
Fang Y:
Circular RNAs as novel biomarkers with regulatory potency in human diseases.
Future Science OA.
2018; 4(7): FSO314. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 73.
Cortés-López M, Gruner MR, Cooper DA, et al.:
Global accumulation of circRNAs during aging in Caenorhabditis elegans.
BMC Genomics.
2018; 19(1): 8. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 74.
Gruner H, Cortés-López M, Cooper DA, et al.:
CircRNA accumulation in the aging mouse brain.
Sci Rep.
2016; 6: 38907. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 75.
Westholm JO, Miura P, Olson S, et al.:
Genome-wide Analysis of Drosophila Circular RNAs Reveals Their Structural and Sequence Properties and Age-Dependent Neural Accumulation.
Cell Rep.
2014; 9(5): 1966–80. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 76.
Jamal M, Song T, Chen B, et al.:
Recent Progress on Circular RNA Research in Acute Myeloid Leukemia.
Front Oncol.
2019; 9: 1108. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 77.
Handschuh L:
Not Only Mutations Matter: Molecular Picture of Acute Myeloid Leukemia Emerging from Transcriptome Studies.
J Oncol.
2019; 2019: 7239206. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 78.
Liu Y, Cheng Z, Pang Y, et al.:
Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia.
J Hematol Oncol.
2019; 12(1): 51. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 79.
Mei M, Wang Y, Li Z, et al.:
Role of circular RNA in hematological malignancies.
Oncol Lett.
2019; 18(5): 4385–92. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 80.
Guo S-S, Li B-X, Zou D-B, et al.:
Tip of the iceberg: Roles of circRNAs in hematological malignancies.
Am J Cancer Res.
2020; 10(2): 367–82. PubMed Abstract
| Free Full Text
- 81.
Naeli P, Pourhanifeh MH, Karimzadeh MR, et al.:
Circular RNAs and gastrointestinal cancers: Epigenetic regulators with a prognostic and therapeutic role.
Crit Rev Oncol Hematol.
2020; 145: 102854. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 82.
Sun J, Li B, Shu C, et al.:
Functions and clinical significance of circular RNAs in glioma.
Mol Cancer.
2020; 19(1): 34. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 83.
Hao Z, Hu S, Liu Z, et al.:
Circular RNAs: Functions and Prospects in Glioma.
J Mol Neurosci.
2019; 67(1): 72–81. PubMed Abstract
| Publisher Full Text
- 84.
Sheng R, Li X, Wang Z, et al.:
Circular RNAs and their emerging roles as diagnostic and prognostic biomarkers in ovarian cancer.
Cancer Lett.
2020; 473: 139–47. PubMed Abstract
| Publisher Full Text
- 85.
Zhang C, Ma L, Niu Y, et al.:
Circular RNA in Lung Cancer Research: Biogenesis, Functions, and Roles.
Int J Biol Sci.
2020; 16(5): 803–14. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 86.
Jahani S, Nazeri E, Majidzadeh-A K, et al.:
Circular RNA; a new biomarker for breast cancer: A systematic review.
J Cell Physiol.
2020; 235(7–8): 5501–10. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 87.
Chaichian S, Shafabakhsh R, Mirhashemi SM, et al.:
Circular RNAs: A novel biomarker for cervical cancer.
J Cell Physiol.
2019; 235(2): 718–24. PubMed Abstract
| Publisher Full Text
- 88.
Jiang P-C, Bu S-R:
Clinical value of circular RNAs and autophagy-related miRNAs in the diagnosis and treatment of pancreatic cancer.
Hepatobiliary Pancreat Dis Int.
2019; 18(6): 511–6. PubMed Abstract
| Publisher Full Text
- 89.
Hua JT, Chen S, He HH:
Landscape of Noncoding RNA in Prostate Cancer.
Trends Genet.
2019; 35(11): 840–51. PubMed Abstract
| Publisher Full Text
- 90.
Hao Q, Han Y, Xia W, et al.:
Systematic Review and Meta-Analysis of the Utility of Circular RNAs as Biomarkers of Hepatocellular Carcinoma.
Can J Gastroenterol Hepatol.
2019; 2019: 684039. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 91.
Zhang Q, Wang W, Zhou Q, et al.:
Roles of circRNAs in the tumour microenvironment.
Mol Cancer.
2020; 19(1): 14. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 92.
Wang Y, Liu J, Ma J, et al.:
Exosomal circRNAs: Biogenesis, effect and application in human diseases.
Mol Cancer.
2019; 18(1): 116. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 93.
Hosaka T, Yamashita T, Tamaoka A, et al.:
Extracellular RNAs as Biomarkers of Sporadic Amyotrophic Lateral Sclerosis and Other Neurodegenerative Diseases.
Int J Mol Sci.
2019; 20(13): 3148. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 94.
Di Liegro CM, Schiera G, Di Liegro I:
Extracellular Vesicle-Associated RNA as a Carrier of Epigenetic Information.
Genes (Basel).
2017; 8(10): 240. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 95.
Zhao H, Chen S, Fu Q:
Exosomes from CD133+ cells carrying circ-ABCC1 mediate cell stemness and metastasis in colorectal cancer.
J Cell Biochem.
2020; 121(5–6): 3286–97. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 96.
Zheng Y, Li JX, Chen CJ, et al.:
Extracellular vesicle-derived circ_SLC19A1 promotes prostate cancer cell growth and invasion through the miR-497/septin 2 pathway.
Cell Biol Int.
2020; 44(4): 1037–45. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 97.
Wang X, Zhang H, Yang H, et al.:
Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer.
Mol Oncol.
2020; 14(3): 539–55. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 98.
Lai RW, Lu R, Danthi PS, et al.:
Multi-level remodeling of transcriptional landscapes in aging and longevity.
BMB Rep.
2019; 52(1): 86–108. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 99.
Li D, Li Z, Yang Y, et al.:
Circular RNAs as biomarkers and therapeutic targets in environmental chemical exposure-related diseases.
Environ Res.
2020; 180: 108825. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 100.
Müller S, Appel B:
In vitro circularization of RNA.
RNA Biol.
2016; 14(8): 1018–27. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 101.
Li J, Mohammed-Elsabagh M, Paczkowski F, et al.:
Circular Nucleic Acids: Discovery, Functions and Applications.
Chembiochem.
2020; 21(11): 1547–66. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 102.
Tatomer DC, Liang D, Wilusz JE:
Inducible Expression of Eukaryotic Circular RNAs from Plasmids.
Methods Mol Biol.
2017; 1648: 143–54. PubMed Abstract
| Publisher Full Text
- 103.
Umekage S, Kikuchi Y:
In vitro and in vivo production and purification of circular RNA aptamer.
J Biotechnol.
2009; 139(4): 265–72. PubMed Abstract
| Publisher Full Text
- 104.
Wesselhoeft RA, Kowalski PS, Anderson DG:
Engineering circular RNA for potent and stable translation in eukaryotic cells.
Nat Commun.
2018; 9(1): 2629. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 105.
Litke JL, Jaffrey SR:
Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts.
Nat Biotechnol.
2019; 37(6): 667–75. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 106.
Umekage S, Tomoe U, Fujita Y, et al.:
In Vivo Circular RNA Expression by the Permuted Intron-Exon Method. INTECH Open Access Publisher: 2012. Publisher Full Text
- 107.
Wang Y, Wang Z:
Efficient backsplicing produces translatable circular mRNAs.
RNA.
2015; 21(2): 172–9. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 108.
Noto JJ, Schmidt CA, Matera AG:
Engineering and expressing circular RNAs via tRNA splicing.
RNA Biol.
2016; 14(8): 978–84. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 109.
Schmidt CA, Noto JJ, Filonov GS, et al.:
A Method for Expressing and Imaging Abundant, Stable, Circular RNAs In Vivo Using tRNA Splicing.
Meth Enzymol.
2016; 572: 215–36. PubMed Abstract
| Publisher Full Text
- 110.
Borchardt EK, Meganck RM, Vincent HA, et al.:
Inducing circular RNA formation using the CRISPR endoribonuclease Csy4.
RNA.
2017; 23(5): 619–27. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 111.
Franz A, Rabien A, Stephan C, et al.:
Circular RNAs: A new class of biomarkers as a rising interest in laboratory medicine.
Clin Chem Lab Med.
2018; 56(12): 1992–2003. PubMed Abstract
| Publisher Full Text
- 112.
Holdt LM, Kohlmaier A, Teupser D:
Circular RNAs as Therapeutic Agents and Targets.
Front Physiol.
2018; 9: 1262. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 113.
Li Z, Ruan Y, Zhang H, et al.:
Tumor-suppressive circular RNAs: Mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets.
Cancer Sci.
2019; 110(12): 3630–8. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 114.
Jost I, Shalamova LA, Gerresheim GK, et al.:
Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges.
RNA Biol.
2018; 15(8): 1032–9. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 115.
Meganck RM, Borchardt EK, Castellanos Rivera RM, et al.:
Tissue-Dependent Expression and Translation of Circular RNAs with Recombinant AAV Vectors In Vivo.
Mol Ther Nucleic Acids.
2018; 13: 89–98. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation


Comments on this article Comments (0)