Keywords
Natural Language Processing, Biomedical Relationship Extraction, NLP, ChemProt, Drug Drug Interactions, Semeval 2010 Task 8
Natural Language Processing, Biomedical Relationship Extraction, NLP, ChemProt, Drug Drug Interactions, Semeval 2010 Task 8
The biomedical literature is a vast corpus of unstructured facts and findings, which need to be synthesised in some systematic way in order for drug discovery scientists to make informed, logical choices about what directions and experiments to pursue. A highly valued goal of biomedical natural language processing (NLP) is to perform relationship extraction (RE) between entities of interest1, such that the knowledge entombed within the literature can be exploited by technological solutions, such as knowledgebase representations. In recent years, groups such as BioCreative and SemEval have coalesced the community around shared RE tasks, in order that we might benchmark our methods against common standards.
Since the early forays into transfer learning to the advent of transformer based models2,3, language modelling and, more recently, masked language modelling is the de rigour methodology in current NLP research. From investigations into the optimal learning objective, to explorations into the limit of pretraining, to permuting the classification head, a bewildering array of research has rapidly emerged, concerning almost every aspect of language modelling. This has created a vast experimental space for the community to explore how such developments relate to biomedical NLP.
The seminal masked language model, Bidirectional Encoder Representations from Transformers (BERT)4, helped to popularise the idea of pretraining on general linguistic data and subsequently fine tuning to tailor the model to downstream tasks. Pretraining is the task of learning some representation of language, such that a piece of text can be encoded into high dimensional space, representing some knowledge about how the tokens within such text relate to each other. Offshoots of BERT, such as SciBERT5, BioBERT6 and BlueBERT7, demonstrated that pretraining on scientific literature allow for better representations of the scientific sublanguage, leading to performance increases in downstream tasks pertaining to that domain. Work such as RoBERTa8 and T59 further recognised that BERT had been undertrained and built upon the original architecture with an expanded pretraining procedure and a larger parameter space.
Although performance gains from larger models and lengthier pretraining is an interesting phenomenon, this represents practical issues for those working within niche domains who desire models pretrained on specific styles of document. With the rapid evolution of new architectures, and substantial costs involved in pretraining, the investment in performing domain specific pretraining becomes hard to justify when the end result may be obsolete within months. Thus, it is desirable to know whether the performance gains from domain specific pretraining outlive the original model architecture (compared to newer architectures that do not benefit from learning better representations of a domain, but perhaps benefit from learning better representations of domain independent, fundamental aspects of language).
A second aspect of language modelling concerns how model are fine-tuned to perform certain tasks. For instance, sentence classification tasks with the original BERT model is possible by passing the sentence representation token (denoted [CLS]) through a linear layer. More recent work (specific to the task of relationship extraction) has explored how combining embedded entity information with such sentence representations can lead to significant performance boosts (the RBERT head)10. However, evidence has since emerged11 that at least some of the perceived performance gains of transformer style models is due to so-called ‘Clever Hans’ type effects, where the model is fine-tuned to learn unintended correlations in datasets rather than a generalised representation of the task. This in turn raises questions about the validity of such approaches in the task of relationship extraction, and how to manufacture appropriate datasets.
The goal of this article is to attempt to address some of these questions via ablation studies of a range of popular masked language models and classification heads, to determine their performance on the task of biological relationship extraction.
We experiment with the general purpose pretrained BERT, model, the biomedical domain specific pretrained model, BioBERT, and the more recent general purpose RoBERTa model. Both BioBERT and RoBERTa are particularly relevant to the ablation tests in this study and serve as an example of a domain-specific model, and a larger model that has undergone lengthier pretraining, respectively. We combine these pretrained models with two classification heads; the commonly used linear layer based on the sentence vector produced by the final layer, and the RBERT classification head. In addition, we examine the effect of four string preprocessing techniques (two per classification head), to investigate how the differing transformer architectures respond to ablations.
We consider ablations over three different corpora labelled with named entities and relationships. The ChemProt12 dataset was originally created for the BioCreative VI workshop, and sought to challenge teams to deliver systems that extracted chemical protein relationships from the scientific literature. It consists of a set of 15,739 relationships annotations from 1,682 PubMed abstracts, divided into training, development and evaluation sets. The dataset covers 11 different label types, although only five undirected relationship types are used in the official evaluation. An official evaluation script is provided.
The DDI (Drug/Drug Interaction) corpus (hereafter DDI) was created for the SemEval-2013 DDI Extraction 2013 challenge13, and seeks to provide a dataset to support the development of NLP systems to extract various types of drug/drug interaction. It consists of 5,028 sentence level relationships manually annotated from Medline and DrugBank, labelled with one of five undirected classes (four describing different types of interaction and one null relationship class) and split into training and evaluation sets. The distribution of labels in this corpus is heavily weighted towards null relations, and thus the imbalance of classes represents an interesting problem for ML classifiers in its own right. The provided official evaluation script calculates the macro F1 over four relationship classes (the null relation is not considered).
Finally, we make use of the Semeval 2010 Task 8 corpus14 (hereafter Semeval), which is a general English RE dataset collected from the Web, and uses a more abstract relationship classification schema than ChemProt or DDI. Here, ten relationship classes are used to annotate 10,717 sentences, which are split into a training set of 8,000 and an evaluation set of 2,717. The provided official evaluation script calculates the macro F1 over nine of the classes in a bi-directional fashion for a total of 18 classes.
For consistency with existing literature, we report our official scores (using official evaluation scripts provided with each dataset) but focus our analysis around cross validation on each set, in order to assess the consistency of the corpora and the effect of random seeds. Here, we report the mean macro averaged F1 score with a five-fold cross validation split.
Originally, we planned to conduct an analysis comparing a wide range of transformer architectures. However, our preliminary investigations suggested that many were too cumbersome to work with, either in terms of compute required, the quality of the pretrained model or the maturity of the codebase. To this end, we restricted our analysis to the pretrained models BERT Base, BioBERT 1.1, RoBERTa base and RoBERTa large, as described in Table 1. Our principal question compares the evaluation performance of BERT Base, BioBERT, and RoBERTa base, as models of approximately equal parameter counts. However, we additionally decided to include RoBERTa large to explore any potential benefits from using a larger model with a higher quality pretraining regime (based upon General Language Understanding Evaluation benchmark results15).
All experiments were conducted with the HuggingFace Transformers implementations, version 2.4.116
Pretrained models are frequently employed in classification tasks, wherein a linear layer is constructed on top of the final layer. Recently, some modifications of this approach have been proposed, to combine specific entity information into the classification layer, to support relationship classification tasks. Wu and He10 suggested averaging the token pieces representing each entity, and concatenating the output with the sentence vector before applying a fully connected feed forward layer, giving rise to the RBERT classification head and setting a new benchmark in the Semeval 2010 Task 8 dataset. In this work, we compare both the simple linear layer classification head and the RBERT head.
RE is commonly construed as a sentence classification task, wherein the label assigned to the relationship between two entities in a sentence are instead assigned to the sentence. However, such an approach can be problematic; for instance, if there are more than two entities in a sentence, and/or more than two relationships (a common occurrence in biomedical text), leading to a situation where the same sentence can yield two conflicting labels.
To mitigate this, various strategies have been used, such as substituting the entities of interest with nominal placeholder tokens, such that all strings seen by a classifier are unique and creating the possibility for a classifier to learn the syntactic importance of the placeholder tokens with regard to the relationship that binds them17. In contrast, the RBERT architecture depends on inserting special characters around the two entities of interest, to inform the classifier of the two input entities without removing information about the entity itself.
Here, we employ ablations on these preprocessing strategies depending on the type of classification head used with the pretrained model (Table 2).
The purpose of the sentence splitting ablation is to provide a baseline classification performance for the underlying pretrained model, without any special characteristics applied to the entities of interest (note, all other transformations include this sentence splitting step). The placeholder transformation is a commonly used strategy in RE6,18,19, where the entities in question are masked by some arbitrary token, thereby attempting to reduce overfitting of the classifier and allowing different relationships between different entity pairs in the same sentence to be represented. Similarly, the bounding special characters ablation is the original transformation as described in the original RBERT paper, whereas the purpose of the masked bounding special character transformation is to remove any entity specific information from the RBERT head. By removing this entity information, our intent is to explore the extent to which the positional information of the entity pairs are used in making the relationship classification, as opposed to the entity embedding information of the entity pairs.
Since some of the preprocessing strategies can lead to undesirable mutations of the underlying data (for instance, it is not possible to represent discontinuous entity boundaries, or overlapping entity boundaries for the placeholder or bounding special character strategies), we filter out any such instances that cannot be transformed for all pretrained model/classification head configurations, such that our training and evaluation sets are consistent across all experiments.
In this ablation study, we aspire for consistency across experiments, rather than attempting to optimise for overall evaluation performance across our selected datasets. To this end, we do not attempt a hyper parameter search. Instead, we defer to recommended hyperparameters for classification tasks based upon the General Language Understanding Evaluation benchmark, as described in the original BERT and RoBERTa papers (Table 3).
One important consideration in hyperparameter selection is the maximum sequence length used. Naturally, it is desirable to use a sequence length big enough to enable the longest sentence in each dataset to be passed though the model. However, longer sequence lengths rapidly increase the memory usage in GPUs, and thus a variable batch size must be selected as required for a given dataset. Since larger batch sizes tend to be desirable20, we originally sought to specify a minimum batch size of 16 across all experiments, in line with recommendations in the BERT and ROBERTA papers. However, initial experiments uncovered that larger models such as RoBERTa large were unable to handle the required sequence length and batch size on the hardware available to us (Tesla V100 16Gb GPUs). To overcome this, we reduced the batch size to four and used eight gradient accumulation steps in all experiments.
We executed six runs across each dataset, per experiment configuration. The first run used the official train/test splits as described in the original datasets, whereas the remaining five runs were comprised of cross validation runs, varying the random seed between folds.
We trained for a maximum of five epochs, and after the first epoch, implemented an early stopping regime that tested for improvements in the average micro F1 score across all classes, after every 5% of the dataset. Five successive failures to improve the F1 resulted in the termination of training, and we logged the highest macro F1 scores reached during training for our cross validation results.
The results of each of our ablations are presented in Figure 1 (tabularised in Table 4).
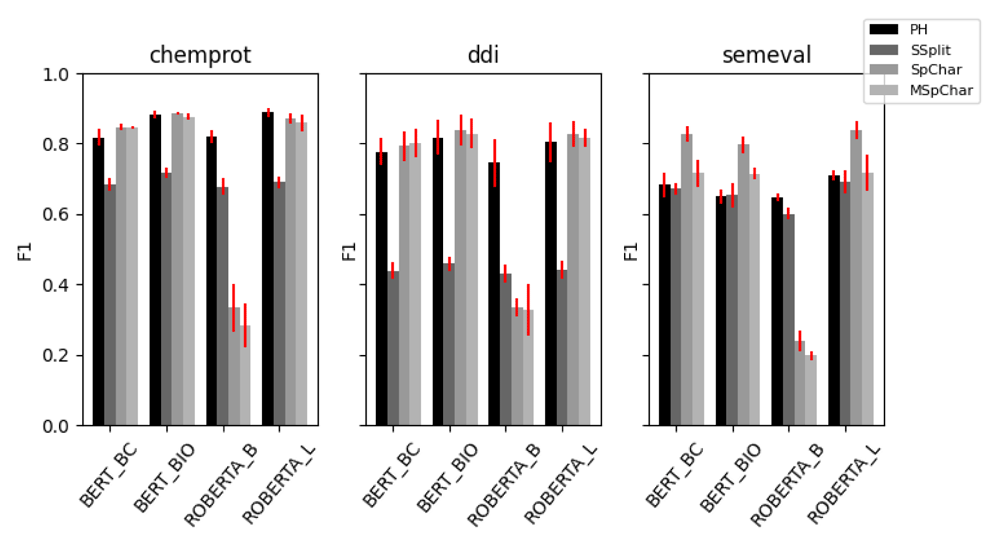
BERT_BC = BERT base cased, BERT_BIO = bioBERT, ROBERTA_B = RoBERTa base, ROBERTA_L = RoBERTa large, PH = placeholder, SSplit = sentence splitter, SpChar = bounding special characters, MSpChar = masked bounding special characters.
BERT_BC = BERT base cased, BERT_BIO = bioBERT, ROBERTA_B = RoBERTa base, ROBERTA_L = RoBERTa large, PH = placeholder, SSplit = sentence splitter, SpChar = bounding special characters, MSpChar = masked bounding special characters. F1 values represents macro F1. Std = standard deviation for cross validation. Random token results use randomly ordered tokens in training data (evaluation data is kept intact).
With respect to the differences with BERT base cased and BioBERT, we observe a moderate benefit by using the BioBERT model on the biomedical Chemprot and DDI datasets, and a moderate benefit by using BERT base on SemEval, in line with observations that domain specific training can improve performance. However, BioBERT and RoBERTa large appear to be approximately equivalent across all datasets, with RoBERTa large ranking marginally higher in most experiments. The surprisingly poor performance of the RoBERTa base model compared to BERT base suggests that most of RoBERTa large’s performance is due to the higher parameter count, rather than the larger size of RoBERTa’s pretraining corpora. Nevertheless, given the very poor performance of the RoBERTa base model with the RBERT head, we are unable to rule out other factors. Particularly difficult to separate is the benefit of training on domain specific data; although RoBERTa is not a biomedical specific model, we examined the contents of the OpenWebText corpora upon which it is trained, and discovered over 11,000 references to PubMed abstracts, as well as other references to providers of scientific literature, suggesting that some of RoBERTa’s performance on biomedical text may come from partial exposure to the domain during pretraining.
On the biomedical datasets, the RBERT classification head seems to provide a small benefit in the DDI task, but this is not observed on the classification performance of ChemProt compared to the placeholder string transformation with the linear layer classifier head. However, the RBERT head appears to substantially boost performance on the SemEval dataset, although the benefits are substantially reduced if entity information is masked. In the case of SemEval dataset, this suggests that the classifier is making more use of the contextual entity embedding than the positional information of the token, and is therefore reliant on latent correlations between the entity pairs and the label, rather than an interpretation of the syntax of the sentence. In the case of the biomedical datasets, many of the classification head/string transformations performed similarly, suggesting none of these are particularly important and that the attention mechanism itself is mostly responsible for learning a representation of the data. A potentially related finding from our results is that even simple sentence classifiers give reasonable performance on the ChemProt and SemEval datasets, with no knowledge of what entity pairs in a sentence the label refers to. To explore this further, we randomised the token order for each instance in the training sets and repeated our experiments for the sentence splitter and placeholder string transformations (Figure 2). Although this ablation created a marked drop in performance across all datasets, we were surprised that this drop was not as substantial as we might have expected. By removing all syntactic information from the training data, it would appear (to a varying degree) that the classifiers are still able to learn some aspects of the relationship classification task using only contextualised embedding information.
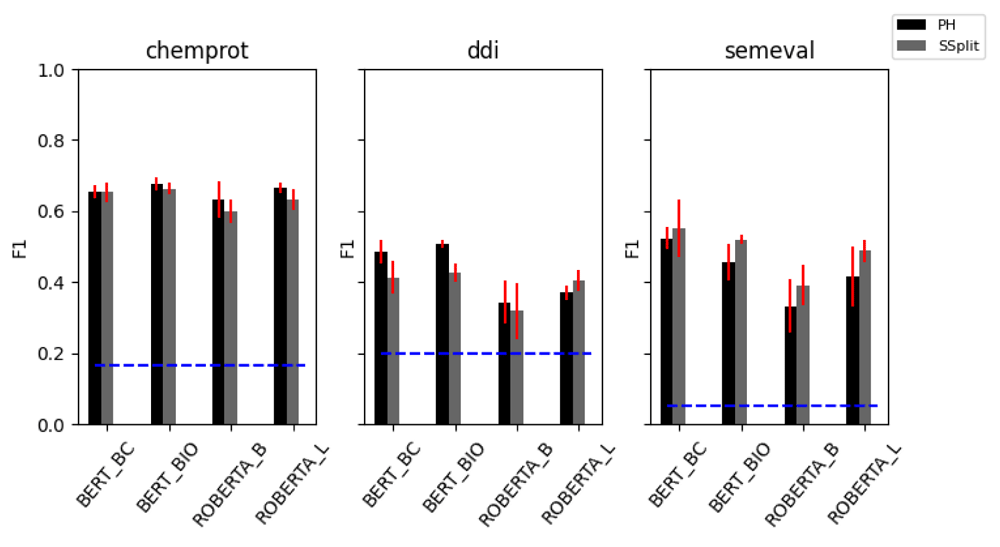
BERT_BC = BERT base cased, BERT_BIO = bioBERT, ROBERTA_B = RoBERTa base, ROBERTA_L = RoBERTa large, PH = placeholder, SSplit = sentence splitter. The horizontal blue line indicates the expected performance of a random classifier.
We suspect that this effect is likely to be attributed to the nature of the underlying training data. Although the attention mechanism employed by the models we tested should be able to learn the required syntactic relationships in order to perform the RE task21, it is also possible for them to learn other aspects of the training data that correlate the sentence embedding information with the given label. For instance, it seems likely that the the presence of certain words may occur more frequently with certain label types, such as verbs suggesting gene regulation activities in the case of ChemProt. Such an effect has recently been established for various NLP architecture across natural language inference datasets, including BERT22. Therefore, models that are trained to make use of such non-syntactic information probably generalise poorly, although further work will be required to establish this conclusively.
In this study, we perform a variety of ablations over an array of models and configurations over three RE datasets. We find that there are benefits in using models pretrained on biomedical text, but the benefits tend to be relatively small and/or task specific on the datasets we explored. Further, there is a tendency of newer models to be trained on larger corpora of text, which appear to encompass the biomedical domain. Future work might revisit analyses such as ours, to determine whether the benefits of domain specific model training outweigh the costs. Finally, we suggest that care must be taken in the training of models for RE, as it appears likely that classifiers are susceptible to overfitting on non-syntactic features. This may be alleviated by the creation of training data that depend heavily on syntactic features, and advancing other methodologies such as data augmentation and Universal Adversarial Triggers23.
The DDI dataset is available from https://github.com/isegura/DDICorpus.
The ChemProt dataset is available from https://biocreative.bioinformatics.udel.edu/news/corpora/chemprot-corpus-biocreative-vi/.
The Semeval 2010 task 8 dataset is available from http://semeval2.fbk.eu/semeval2.php?location=data.
Source code available from: https://github.com/RichJackson/pytorch-transformers
Archived source code at time of publication: http://www.doi.org/10.5281/zenodo.389462524
License: Apache 2.0
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Hoyt C, Domingo-Fernández D, Hofmann-Apitius M: BEL Commons: an environment for exploration and analysis of networks encoded in Biological Expression Language. Database. 2018; 2018. Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Biomedical natural language processing
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Arora: A Simple but Tough-to-Beat Baseline for Sentence Embeddings. 5th International Conference on Learning Representations, ICLR 2017. 2017. Reference SourceCompeting Interests: KV receives funding from Elsevier BV for research on methods for information extraction from biochemical texts and has a related provisional patent.
Reviewer Expertise: Biomedical natural language processing
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 16 Jul 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)