Keywords
Interoperability, eCRF, iCRF, Codebook, FAIR, Software, EDC, Clinical data
Interoperability, eCRF, iCRF, Codebook, FAIR, Software, EDC, Clinical data
Clinical data is essential for health research. Traditionally, such data was captured using paper case report forms (CRFs) and entered into a database manually. Nowadays, the data is often captured directly with electronic CRFs (eCRFs) in an electronic data capture (EDC) system. This has improved the quality of the captured data as well as decreased costs for data collection (e.g. 1,2).
To allow the captured data to be used beyond its original purpose requires the data to be FAIR (Findable, Accessible, Interoperable and Reusable)3. By making data semantically interoperable, it can be exchanged between systems whilst preserving the meaning of the data4. Furthermore, it allows multiple data sources to be combined and understood by computers, thereby e.g. facilitating clinical decision support systems5. Hence, when setting up a new data collection protocol, the eCRF should be designed with interoperability in mind. Achieving semantic interoperability requires the use of a communication standard, such as HL7, as well as (functional) information standards4, such as the NCI thesaurus or SNOMED CT. However, mapping study-specific terminology to a thesaurus requires expert knowledge of the thesaurus, the data and its context. Therefore, reusing existing codebooks from studies and well-known datasets or CRF templates, such as available from CDISC’s CDASH, the Meta Data Models website and the University of Wisconsin-Madison, can be a viable alternative. Reusing these elements at the very minimum facilitates interoperability with other datasets using these definitions. Furthermore, in many cases well-known codebooks have already been mapped to a thesaurus. For example, in the Basic Health Data Set, which is the standard that will be used by hospitals to exchange healthcare data in the Netherlands (available here, Dutch only), many of the items have been mapped to SNOMED CT.
In this paper, we introduce the iCRF Generator, a program that allows users to easily generate interoperable electronic case report forms (iCRFs) based on online codebooks, thereby improving the interoperability of clinical data collected in and between EDCs. Whereas normally CRF generation is an integrated part of the EDC (e.g. Castor EDC, REDCap), our program can generate the core of a CRF for multiple EDCs. At this time, three systems are supported: Castor, OpenClinica 3 and REDCap. The program allows a user to select one or more codebooks available from an online system called ART-DECOR which allows, amongst others, the storage of dataset definitions and select items of interest, including their codelists. The program currently supports six codebooks, which are further described in the Methods section.
The iCRF Generator was written in Java 8 and later migrated to Java 12 for JavaFX compatibility. Dependencies are managed using Maven and include: JavaFX and ControlsFX for the UI, Apache POI for Excel file management and Log4j for logging. A ZIP file of the iCRF Generator distribution is available for both Mac and Windows. It includes a Java Runtime Environment to ensure independence of the locally installed Java version and ensures the program works out of the box. Source and distribution files are available on GitHub: https://github.com/aderidder/iCRFGenerator/.
The iCRF Generator is designed to use codebooks defined in ART-DECOR. ART-DECOR is an open-source tool suite that supports the creation and maintenance of HL7 templates and allows the storage of dataset definitions. Nictiz, the centre of expertise for eHealth and the Dutch SNOMED-CT release centre, facilitates ART-DECOR to create health information standards that are publicly accessible. The iCRF Generator currently offers access to six of these codebooks, which were chosen because of their national relevance (codebooks 1, 2 and 3) and our involvement (4, 5 and 6):
1. The Clinical Building Blocks (Zorginformatiebouwstenen): information models of minimal clinical concepts. They are used as the basis for the Basic Health Data Set.
2. The Basic Health Data Set (Basisgegevensset Zorg): codebook used for the standardised exchange of patient data between e.g. healthcare providers. Implementation of this set is prioritised in healthcare systems like electronic health records. The Basic Health Data Set is aligned with the European Patient Summary.
3. The National Institute for Public Health and the Environment’s national screening codebook of bowel cancer and cervical cancer (RIVM bevolkingsonderzoeken).
4. Cancer Core Europe: a European cancer research alliance which aims at bringing together the expertise and critical mass necessary to make translational research available in the clinic.
5. The PALGA Colon biopsy protocol: PALGA is the nationwide network and registry of histo- and cytopathology in the Netherlands.
Some of the codebooks are available in English, as well as Dutch. Other codebooks may be made available in the iCRF Generator in the future when they become available in ART-DECOR provided they are of sufficient quality and are complementary.
A standard PC or Mac should be able to run the program without any issues. The program was tested on a Windows 7 PC, a Windows 10 PC and on a virtual machine with OS X El Capitan. To give an indication of the program’s memory usage: selecting a single codebook, the program uses around 230 megabytes of memory; increasing this number to eight codebooks increased the memory usage to 420 megabytes. An internet connection is required, as the program retrieves metadata as well as codebooks from ART-DECOR via the REST API.
A typical use case for the iCRF Generator is in the design phase of a study or registry. When a decision has been made on what clinical data is going to be collected, the data manager has to design and build the CRFs for data collection. Instead of doing this manually, the iCRF Generator can be used to select items from the available codebooks and generate the basis of the case report forms. Figure 1 illustrates the iCRF Generator’s complete workflow. In a typical use case, the user first selects an EDC from the dropdown (Figure 2). The user then clicks the “Run” button, after which the wizard interface is started. The first wizard page asks the user to select one or more codebooks (Figure 3). When a user has selected a codebook and presses the “Next” button, a REST-call is made to retrieve an XML which contains which versions and languages are available for the codebook. The second page shows this information to the user, allowing selection of the versions and languages of interest (Figure 4). When the user proceeds to the next page, the selected codebooks are retrieved via one or more REST-calls. These XML files are parsed and the information it contains about the items and their possible values is shown on the third page (Figure 5). This EDC-specific page allows users to select and customise the items that have to be included in the CRF. The final page of the wizard shows a short summary of the number of selected items. Upon completion of the wizard, the program generates the CRF in the format required by the selected EDC and the file is saved to disk. This file can then be imported into the EDC-system or opened in an editor of choice.
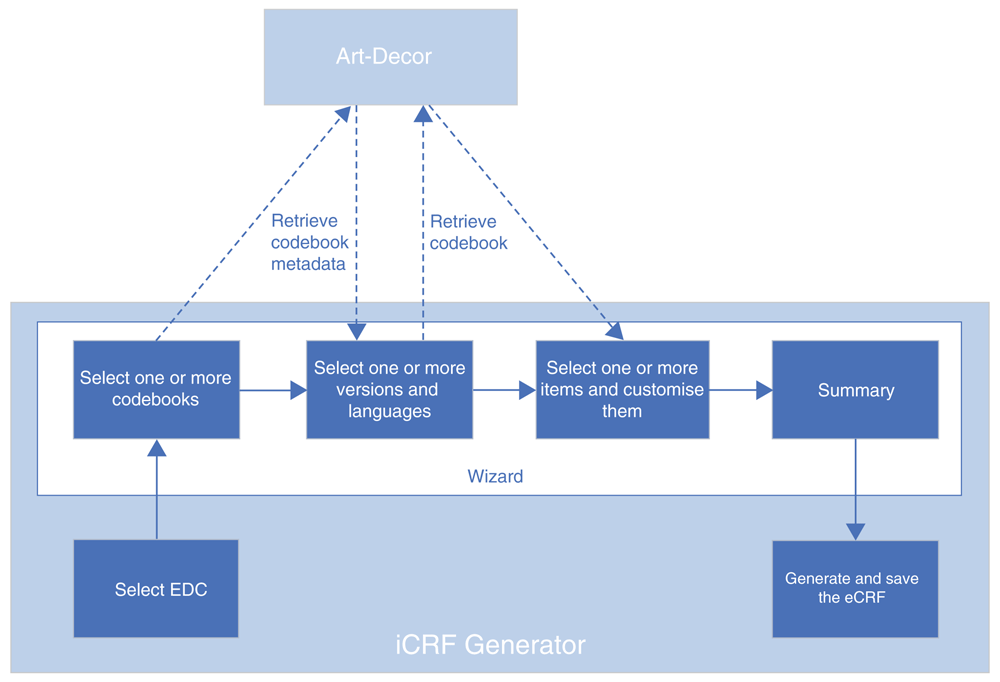
Dotted lines are calls made by the program to ART-DECOR’s REST services.
To allow for the use of data beyond its original purpose, it is essential that the data is FAIR (Findable, Accessible, Interoperable and Reusable)3. To preserve meaning and intent of clinical data when it is exchanged, requires the data to be semantically interoperable, which requires the use of content standards. Manually mapping data definitions to a medical thesaurus such as SNOMED CT is complicated and time consuming. Instead, reusing existing published definitions (codebooks) can be a viable alternative, as interoperability with other datasets using these codebooks is usually easily achieved. Furthermore, the codebooks may already have been mapped to a thesaurus.
In this paper we introduced the iCRF Generator, a program which can generate electronic case report forms for three major electronic data capture systems: Castor, REDCap and OpenClinica 3. The program allows a user to select items and codelists from several highly relevant codebooks available from the online system ART-DECOR. By providing an easy to use program to generate CRFs using these codebooks, data will be collected using the same definitions, which enhances the interoperability and FAIRness of the data.
One important usability aspect of software is the software’s performance. The codebooks available in the iCRF Generator are parsed from XML files generated by ART-DECOR. It takes ART-DECOR around 30 seconds to generate the XML file for the National Institute for Public Health and the Environment screening codebook. Furthermore, in some cases XML files can reference other XML files, which then have to be downloaded and parsed as well. If a user has to wait every time a codebook is selected, user acceptance will quickly erode. Hence, we introduced local caching of downloaded codebooks, which makes usage nearly instantaneous once the codebook is locally available. Furthermore, we intend to make a ZIP file of the cache available for download. Note that downloading a codebook XML from ART-DECOR only takes place if it is accessed for the first time or when a new version of a codebook becomes available and is selected by the user.
The iCRF Generator can easily be expanded to include additional EDCs, such as OpenClinica 4, Research Manager and Alea if there is demand and the import formats are available. If desirable, additional internationally established formats, such as CDISC ODM, may also be included in the future.
At this point, the iCRF Generator gives access to six codebooks, some of which support multiple languages and multiple versions. When more codebooks are published in ART-DECOR, we intend to make these available in the program, provided they are of sufficient quality and are complementary to the codebooks already available. Criteria to decide whether a codebook will be made available will be established in the near future. We believe that providing too many overlapping codebooks will be counterproductive for interoperability as items may well map to different thesauri. Furthermore, the user may be overwhelmed by the available information.
When an item is selected in the item tree, a user can customise the item. As each EDC has different requirements for its CRFs, the customisation options we provide vary per EDC. As an example, OpenClinica 3 has a “Field Type” (e.g. “Radio”, “Single-Select”) and a “Data Type” (e.g. “ST”, “INT”), whereas in Castor the data type does not exist as a separate entity.
The customisation options we currently provide are limited. The iCRF Generator’s purpose is to facilitate generation of interoperable CRFs. Hence, if everything could be customised, for example replacing the codes in codelists with custom codes, it would undermine the purpose of the program. Furthermore, the iCRF Generator is work-in-progress and some further item customisation may be added in the future.
A tool somewhat similar to our own is ODMedit6. ODMedit provides a web-based interface to allow users to create a CRF based on elements stored in the Meta Data Repository. When a user has finished creating the CRF, it can either be downloaded in ODM format or uploaded to the Medical Data Models-portal. From there it can be downloaded in multiple formats.
ODMedit differs from our software in several ways. Whereas ODMedit immediately provides access to all items in its repository, we keep our items grouped by codebook. Furthermore, with ODMedit users can immediately add new and edit existing items, and new items are automatically made available in the repository. In our tool we are providing access to only handpicked codebooks, from which the user can select items and customisation of these items is kept to a bare minimum. By allowing users to select items from well-known and supported codebooks only, we believe it should be easier to find the correct item - e.g. if you need pathology definitions, use items from the pathology codebooks. However, we may have to add a search function at some point to make it easier to find items within a codebook. Another difference is that we decided to explicitly ask for which EDC tool the user wishes to create the CRF to allow for EDC specific options. On the other hand, ODMedit does support some features which we do not yet support, such as a repeating group. We may add this at a future time.
An OpenClinica 3 specific CRF generator is also available. This tool converts a csv file to Excel and provides a user with an interface to edit the CRF. However, the tool does not facilitate interoperability.
Multiple initiatives exist that aim at providing templates to improve interoperability. We list several such initiatives below. The National Institute of Health offers Common Data Elements, data elements that are common to multiple data sets across different studies. CDASH, provided by CDISC, gives guidance for developing CRFs used in clinical trials7. The OpenClinica Building Blocks developed by TraIT provide OpenClinica users with templates to which they can add study-specific items and remove items that are not necessary for their study. The Australian Government launched a platform for digital health. They provide an extensive library of documents, tools and much more for implementers and developers. The Global Alliance for Genomics & Health (GA4GH) has several workstreams, amongst which one for Clinical & Phenotypic Data Capture, that “Supports the clinical adoption of genomics through establishing standard ontologies and information models to describe the clinical phenotype for use in genomic medicine and research, including the capture and exchange of information between electronic clinical systems and research.”
Source code available here: https://github.com/aderidder/iCRFGenerator
Archived source code as at time of publication: https://doi.org/10.5281/zenodo.35635008
License: GNU GPL v3 license.
We thank Jan-Willem Boiten (Lygature), Gerben Rienk Visser (Trial Data Solutions) and Maarten Ligtvoet (Nictiz) for reviewing this paper and providing invaluable suggestions. We also thank Wessel Sloof (UMCG) for testing the generated REDCap exports.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
References
1. Dugas M, Neuhaus P, Meidt A, Doods J, et al.: Portal of medical data models: information infrastructure for medical research and healthcare.Database (Oxford). 2016; 2016. PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Medical Informatics, Medical Data Models, semantic annotations, interoperability
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Partly
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
No
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: Petros Kalendralis (co-reviewer) works on the Trait2HealthRI research project
Reviewer Expertise: Our area of research focuses on making medical data FAIR. For example, we employ the Personal Health Train to share information across medical centers in a privacy-preserving manner.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 23 Mar 20 |
read | read | |
|
Version 1 04 Feb 20 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)