Keywords
COVID-19, SARS-CoV-2, Synonymous mutations, RNA secondary structure
This article is included in the Emerging Diseases and Outbreaks gateway.
This article is included in the Research Synergy Foundation gateway.
This article is included in the Coronavirus (COVID-19) collection.
COVID-19, SARS-CoV-2, Synonymous mutations, RNA secondary structure
In December 2019, coronavirus disease 2019 (COVID-19) cases first emerged from Wuhan, China1. Soon after, rapid spread of COVID-19 has resulted in a serious global outbreak. COVID-19 is an infectious and potentially lethal disease caused by a newly found coronavirus strain, known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The virus causes clinical manifestation ranging from asymptomatic to severe pneumonia and eventually death2. SARS-CoV-2 seems to have a higher transmission rate3 but lower mortality rate2 in comparison to Middle East respiratory coronavirus (MERS-CoV) and severe acute respiratory syndrome coronavirus (SARS-CoV).
SARS-CoV-2 is a single-stranded RNA virus with a genome size of 29,903 bases. In general, RNA viruses have a higher mutation rate than DNA viruses and this allows them to evolve rapidly, escaping the host immune defence response4. A study by Kim et al. (2020) identified a total of 1,352 nonsynonymous and 767 synonymous mutations from 4,254 SARS-CoV-2 genomes5. While in another study done by Khailany et al. (2020), 116 mutations had been identified from 95 SARS-CoV-2 genomes6.
In this study, we focus on synonymous mutations instead of nonsynonymous mutations as researchers often overlook their biological importance. Synonymous mutations are also known as silent mutations because the nucleotide mutations result in a change in the RNA sequence without altering the amino acid sequence7. Synonymous mutations have been suggested to have no functional consequence on the fitness of organisms and their evolution in long term8. However, numerous recent studies had showed that synonymous mutations may affect the folding and stability of RNA structures9. For RNA viruses, even though synonymous mutations generally do not change their pathogenicity, some studies reveal that synonymous mutations may affect the RNA secondary structure of the virus10 and also change the codon usage bias of the genes in the virus11,12.
30,229 SARS-CoV-2 genomic sequences were retrieved from GISAID database (Global Initiative on Sharing All Influenza Data, RRID:SCR_018251)13 ranging from 31 December 2019 to 22 March 2021. SARS-CoV-2 genomic sequences were filtered by setting parameters to keep only sequences with complete genome and high coverage. The reference sequence of SARS-CoV-2 genome (NC_045512.2)14 was retrieved in fasta format from NCBI database (NCBI, RRID:SCR_006472). It is a Wuhan isolate with a complete genome which comprises of 29,903 bases.
The rapid calculation available in MAFFT online server (MAFFT, version 7.467, RRID:SCR_011811)15 was used to perform multiple sequence alignment (MSA) for 30,229 SARS-CoV-2 genomes. This option supports the alignment of more than 20,000 sequences with approximately 30,000 sites. The alignment length was kept, which means the insertions at the mutated sequences were removed, to keep the alignment length the same as the reference sequence. While other parameters were left as default.
A simple Python script was written to identify the mutations in 30,229 SARS-CoV-2 genomes. To determine whether the identified mutations are synonymous or nonsynonymous, MEGA X software, version 10.2.5 build 10210330 (MEGA Software, RRID:SCR_000667)16 was utilized to perform the translation for inspection purposes. The presence of amino acid changes was identified by referring to the genomic position of the nucleotide mutations. Synonymous mutations with the top 10 highest frequencies were generated.
The RNA secondary structure of wild type and mutant sequences were predicted using RNAfold program, version 2.4.18 (Vienna RNA, RRID:SCR_008550) to show how mutations affect RNA secondary structure. The RNA secondary structure prediction was performed using a sequence length of 250 nucleotides upstream and downstream of the mutation site. The minimum free energy (MFE) was also calculated in the RNAfold program for both wild type and mutant sequences to show comparison of the RNA folding stability between them. The comparison on the value of minimum free energy (MFE) is important to indicate whether the mutations affect the folding stability of the respective RNA structure.
To predict how the mutations affect RNA local folding, base pair probability was estimated by utilizing MutaRNA, version 1.3.0 (MutaRNA, RRID:SCR_021723)17. MutaRNA is a web-based tool that allows prediction and visualization of the structure changes induced by a single nucleotide polymorphism (SNP) in an RNA sequence. It includes the base pair probabilities within RNA molecule of both wild type and mutant. The parameters used in MutaRNA were set as default, in which the window size is 200nt and the maximal base pair span is 150nt.
Relative synonymous codon usage (RSCU) represents the ratio of the observed frequency of codons appearing in a gene to the expected frequency under equal codon usage. RSCU is calculated using the formula:
where Xi implies the number of occurrences of codon i and n stands for the number of synonymous codons encoded for that particular amino acid.
A synonymous mutation is a change in the nucleotide that does not cause any changes in the encoded amino acid. Synonymous mutations were previously considered to be less important, but they are now proven to have some effects on RNA folding, RNA stability, miRNA binding and translational efficiency18. Synonymous mutations may have significant effects on the adaptation, virulence, and evolution of RNA viruses19. Another study done also indicated that synonymous mutations have association with more than 50 human diseases such as hemophilia B, tuberculosis (TB), cystic fibrosis (CF), Alzheimer, schizophrenia, chronic hepatitis C and so on20. All these studies show that increasing importance has been associated with synonymous mutations over these years. Hence, it is necessary for us to study the effects of synonymous mutations of SARS-CoV-2 genome.
A total of 381 mutations were found in SARS-CoV-2 genomes by using python script, in which 150 of them are synonymous mutations. The distribution of synonymous mutations in 11 coding regions is shown in Figure 1. Among these mutations, ORF1a and ORF1b have a higher number of synonymous mutations at 76 and 33, respectively, which might be due to their longer sequence length. Besides that, our findings also show high C to U mutation rate in SARS-CoV-2 genetic variation and this mutational skews are in line with the studies done by Rice et al. (2021) and Simmonds (2020)21,22. These mutational skews are necessary to be considered when deducing the selection acting on synonymous variants in SARS-CoV-2 evolution23.
The synonymous mutations in SARS-CoV-2 genomes with the top 10 highest frequency were listed in Table 1. As shown in Table 1, synonymous mutations with the highest frequency identified from SARS-CoV-2 genomes is C3037U mutation located in nsp3 of ORF1a, followed by C313U mutation in nsp1 of ORF1a and C9286U mutation in nsp4 of ORF1a. Mutations with higher frequency are mostly found in ORF1a and ORF1b. It is of great interest to find out the effect of these top 10 synonymous mutations on SARS-CoV-2 genome.
SARS-CoV-2 virus can form highly structured RNA elements, which may affect viral replication, discontinuous transcription and translation24–28
The RNA secondary structures of wild type and mutant sequences were predicted using RNAfold program. Minimum free energy (MFE) of each structure was also calculated to show the folding stability of the respective RNA structure. Among all, C913U mutation and C26735U mutation were found to have a more obvious effect on the predicted RNA secondary structure compared to the wild type.
As shown in Figure 2(A), a single hairpin is formed around the mutant with C913U mutation instead of a multiloop in the wild type. The minimum free energy value of the mutant (- 146.90 kcal/mol) is slightly less negative than that of the wild type (- 147.40 kcal/mol), which makes it a less thermodynamically stable structure compared to the wild type.
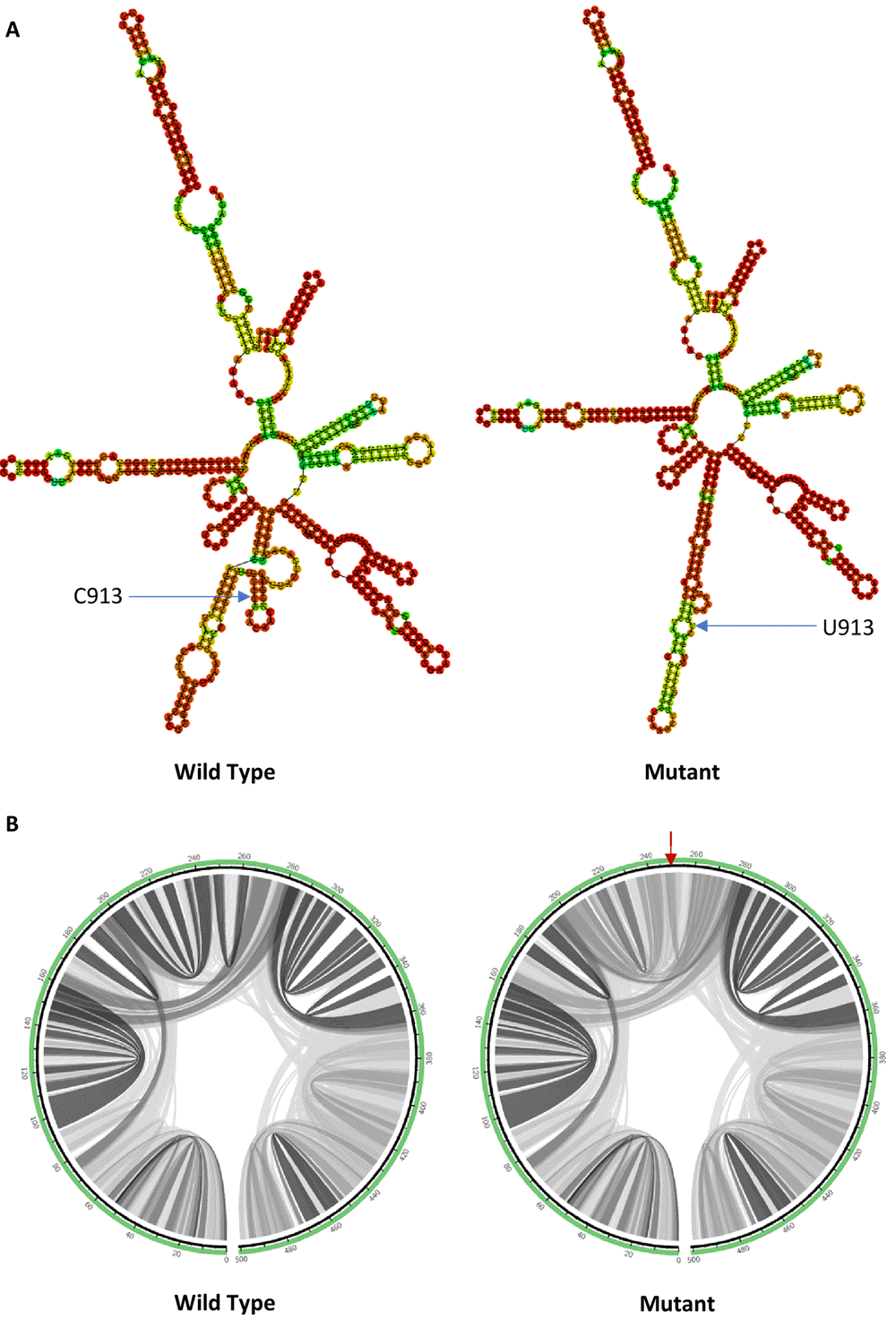
(A) RNA secondary structure of wild type C913 and mutant U913. (B) Circular plots showing the base pairing probabilities with darker shades indicating higher base pairing probabilities and a red arrow pointing to the mutation site.
To visualize the differences in the base pairing potential induced by the mutation, base pairing probability was estimated using MutaRNA. The circular plots in Figure 2(B) show the base pairing probabilities of both wild type and mutant. As shown in the circular plots, C913U mutation decreased the Watson–Crick base pairing probability near the mutation site, which led to a less stable predicted RNA secondary structure. C913U mutation is found in the nsp2 of ORF1a in SARS-CoV-2 genome. Nsp2 in SARS-CoV interacts with two human host proteins specifically, which are prohibitin 1 (PHB1) and prohibitin 2 (PHB2)29. This interaction may disrupt the intracellular host signalling during the viral infections, rather than taking part in the viral replication29. It is yet to see if nsp2 of SARS-CoV-2 shares same or similar function as that of SARS-CoV.
C26735U mutation is located at the membrane (M) protein. M protein may suppress host immune responses via the interference of Type I interferon production30. C26735U mutation induces changes in the predicted RNA secondary structure by forming an extra multibranch loop at the mutation site as shown in Figure 3(A). The RNA secondary structure formed by the mutant (-134.50 kcal/mol) has a less negative minimum free energy value compared to the wild type (-136.30 kcal/mol), which makes it a less stable structure. While for the circular plots in Figure 3(B), C26735U mutation decreases the base pairing probabilities in flanking regions, which again predicted that the mutation decreases the folding stability of the RNA secondary structure.
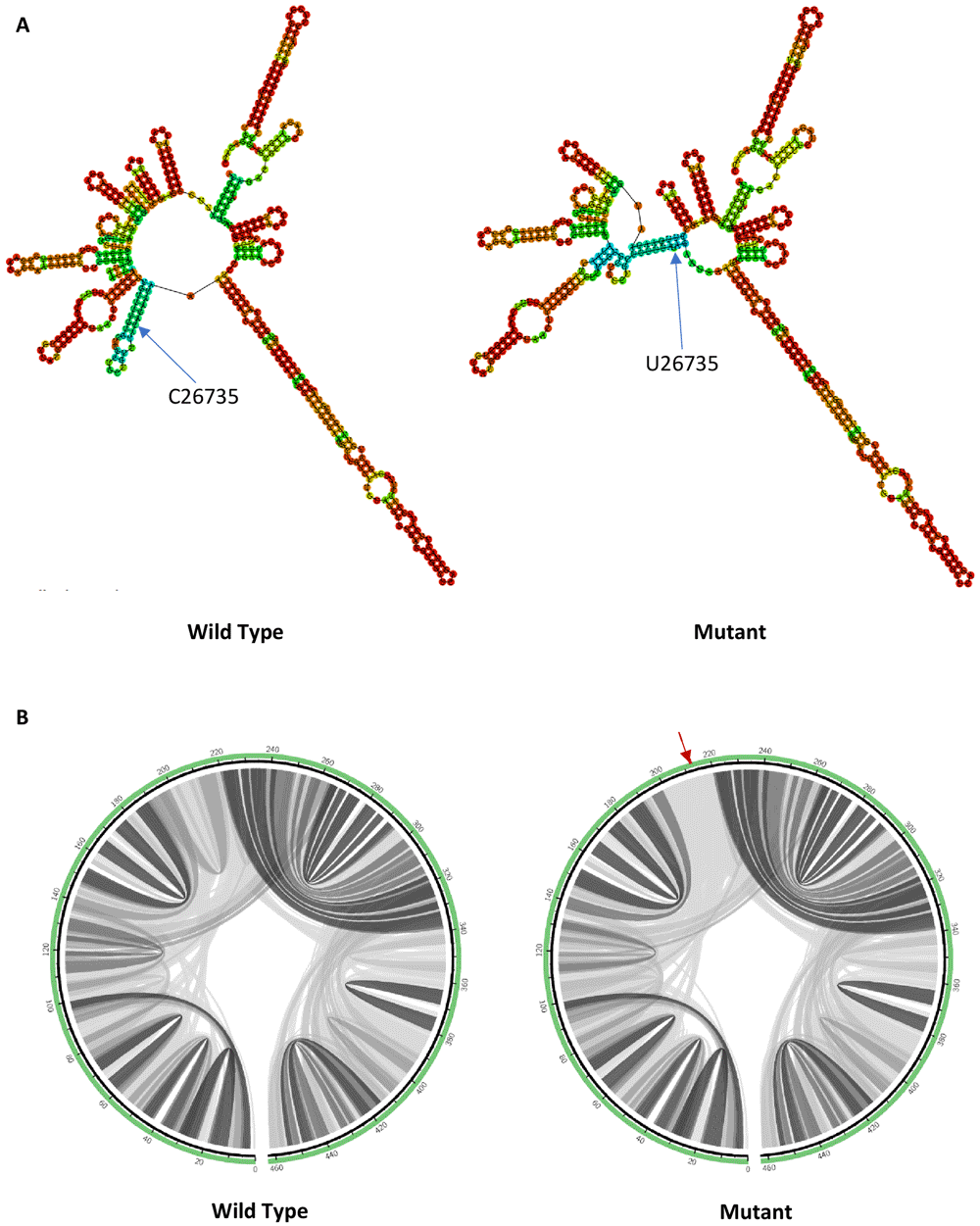
(A) RNA secondary structure of wild type C26735 and mutant U26735. (B) Circular plots showing the base pairing probabilities with darker shades indicating higher base pairing probabilities and a red arrow pointing to the mutation site.
In short, both C913U and C26735U mutations cause a more drastic change in RNA secondary structure. Given the shortcomings of prediction tools, it is necessary to check if the changes in RNA secondary structure affect the pathogenicity of SARS-CoV-2 using experimental approaches. Besides that, these two mutations also reduce the folding stability of the RNA secondary structure, which then affects the polypeptide translation and folding. There is evidence suggesting that stable RNA structures play a key role in reducing the translation speed to prevent “ribosomal traffic jams” so that the newly translated polypeptides can fold properly31. Hence, both C913U and C26735U mutations increase the translation speed of SARS-CoV-2 RNA but they might cause the nascent polypeptide folding more prone to error during translation.
Codon usage bias (CUB), which is non-random usage of synonymous codons, is common in all species. It is a phenomenon where some codons are preferred over others for a specific amino acid. SARS-CoV-2 replicates using host cell’s machinery and synthesizes its protein by utilizing host cellular components. Hence, codon usage bias may affect the replication of viruses32.
Relative synonymous codon usage (RSCU) is a widely used statistical approach33 that can be used to measure codon usage bias in coding sequences. The RSCU values of SARS-CoV-2 are shown in Table 2 and the most preferred codons for each amino acid are marked in bold. Stop codons (UAA, UAG, UGA) and codons which code for an amino acid uniquely (AUG, UGG) are excluded from RSCU analysis.
Based on the RSCU values, the synonymous codons can be classified into five groups: i) codons with RSCU value equals to 1.0 are unbiased codons; ii) codons with RSCU value > 1.0 are codons preferred in a genome; iii) codons with RSCU value < 1.0 are codons less preferred in a genome; iv) codons with RSCU value > 1.6 are codons which are over-represented in a genome; v) codons with RSCU value < 1.6 are codons which are under-represented in a genome32. There are 15 preferred codons (RSCU value > 1.0) and 11 over-represented codons (RSCU value > 1.6) in SARS-CoV-2 genome as shown in Table 2. The preferred codons in SARS-CoV-2 genome are GCA (Ala), CGU (Arg), AAU (Asn), GAU (Asp), UGU (Cys), CAA (Gln), GAA (Glu), CAU (His), AUU (Ile), UUG (Leu), AAA (Lys), UUU (Phe), CCA (Pro), AGU (Ser) and UAU (Tyr) while the over-represented codons are GCU (Ala), AGA (Arg), GGU (Gly), CUU (Leu), UUA (Leu), CCU (Pro), UCA (Ser), UCU (Ser), ACA (Thr), ACU (Thr), and GUU (Val). The presence of the preferred and over-presented codons in a genome increases the protein synthesis rate.
Table 3 shows the RSCU analysis of the top 10 synonymous mutations. The codons in bold in the ‘codon change’ column are the codons with higher RSCU value, which means they are more preferred in SARS-CoV-2 genome. Most of the mutations change the codon to a more preferred codon as shown in Table 3. Since it is presumed that preferred codons have a higher translation rate compared to nonpreferred codons34, it is possible that most of the mutations may increase the translation efficiency of SARS-CoV-2, which may affect virus replication, transmission, and evolution.
The effects of SARS-CoV-2 synonymous mutations in various aspects such as RNA folding and RNA stability of the virus were studied, even though they do not cause changes in amino acid residue of the protein. Most of the synonymous mutations identified in the SARS-CoV-2 genome are found to have a minor effect on RNA folding and RNA stability of the virus except for C913U and C26735U mutations. Due to the shortcomings of prediction tools, experimental studies are needed to give a more comprehensive understanding of the biological consequences of synonymous mutations on SARS-CoV-2 virus.
No ethical approval is required for data analysis in this study (EA2702021).
SARS-CoV-2 virus genome sequence data were obtained from the GISAID Database. The multiple alignment data can be assessed through FigShare.
Figshare: MSA (SARS-CoV-2). https://doi.org/10.6084/m9.figshare.16681900.v135
This project contains the following underlying data.
• MSA_0.fasta (multiple sequence alignment of SARS-CoV-2 sequences obtained between 31-12-2019 and 31-05-2020.)
• MSA_1.fasta (multiple sequence alignment of SARS-CoV-2 sequences obtained between 01-06-2020 and 15-10-2020.)
• MSA_2.fasta (multiple sequence alignment of SARS-CoV-2 sequences obtained between 16-10-2020 and 31-01-2021.)
• MSA_3.fasta (multiple sequence alignment of SARS-CoV-2 sequences obtained between 01-02-2021 to 22-03-2021.)
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
The python code for the identification of SARS-CoV-2 genome mutations can be assessed through GitHub.
CHN contributes to the concept, design, supervision of the project. WXB and SBZ contribute to the design, methodology, and data collection. WXB contributed to the analysis, and interpretation of data.
All authors were involved in drafting and revising the manuscript and approved the final version.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
No
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
No
References
1. Lan T, Allan M, Malsick L, Woo J, et al.: Secondary structural ensembles of the SARS-CoV-2 RNA genome in infected cells. Nature Communications. 2022; 13 (1). Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: RNA structure prediction; Computational Structural Biology; RNA-protein complexes.
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Clinical Virology Diagnosis
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genome Analysis, SARS-CoV-2, Cancer, Immunology, Stem cell
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
|
Version 4 (revision) 18 Sep 24 |
read | |||||
|
Version 3 (revision) 29 Feb 24 |
read | read | ||||
|
Version 2 (revision) 05 Sep 22 |
read | read | ||||
|
Version 1 18 Oct 21 |
read | read | read | |||
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)