Keywords
Bacteriophage genomes, RB49-like bacteriophages, coliphages, O antigens, receptor recognition proteins.
This article is included in the Cell & Molecular Biology gateway.
This article is included in the Genomics and Genetics gateway.
This article is included in the Pathogens gateway.
Bacteriophage genomes, RB49-like bacteriophages, coliphages, O antigens, receptor recognition proteins.
The threat of rapidly spreading microbial resistance to antibacterial drugs has revitalized the interest to the usage of bacteriophages as biocontrol agents for agriculture (Allen et al. 2013), aquaculture (D'Accolti et al. 2021), phage therapy in humans and animals (Abedon 2018, Clokie and Mary Ann Liebert Inc. 2020) and other applications.
One of the main limitations of phage-based biocontrol technologies is narrow host range of most of bacteriophages. T-even-related bacteriophages, including RB49-like viruses, are widely spread in nature and feature significant sequence flexibility in the receptor recognition proteins (RBPs), coupled with an high overall homology of the other regions in their genomes (Comeau et al. 2007). This makes them attractive platforms for developing systems of artificial host range management.
The paradigm of the host cell recognition by T-even-like phages was studied in great detail on the bacteriophage T4 model system. The infection of the host cell by T4 is initiated by binding of the tips of the phage long tail fibers (LTFs) to their cognate receptor molecules (OmpC) (Hu et al. 2015). This initial binding triggers the baseplate rearrangement and deployment of the short tail fibers (Taylor et al. 2016) that bind to different receptors tightly fixing the virion on the cell surface to prevent it from been pushed out by the contraction of the phage tail.
The bioinformatic and experimental evidence suggest that in other T-even-like phages the outline of the early stages of infection is essentially the same as in T4, however in most of these viruses (including RB49 and related phages) the recognition of LTF receptors is performed not by the C-terminal domain of the trimeric gp37 protein forming the distal moiety of the LTF, but by a tiny monomeric protein gp38 attached to the end of the gp37 trimer (Trojet et al. 2011, Hyman and van Raaij 2018).
Interestingly, the above-described host recognition scenario was mainly evaluated by studying the phage T4 infection of the laboratory E. coli strains that are depleted of the O antigen biosynthesis, such are the derivatives of K-12 or B strains (Tetart et al. 2001). At the same time evidence accumulates that many types of E. coli O antigen serve as an effective shields restricting the access of the phage receptor-recognition structures to the receptors, located at the outer membrane surface (Peng et al. 2007, Golomidova et al. 2021) (Broeker and Barbirz 2017). The penetration through this shield requires specific recognition and, in some cases, enzymatic degradation of the O polysaccharide, by the phage RBPs (Letarov and Kulikov 2017, Kulikov et al. 2019) . In order to evaluate the mechanisms employed by T-even-like bacteriophages to attack the O antigen-producing E. coli strains it is necessary to obtain such phages infective against the host strains in which the protective features of the O antigens were studied in some detail. We previously investigated the structure and protectivity of the O antigens of several wild type isolates of E. coli, including the strains 4s (Knirel et al. 2015, Kulikov et al. 2017), F5 (Golomidova et al. 2019), and F17 (Knirel et al. 2019). Here we report the isolation and sequencing of three RB49-like bacteriophages able to infect these strains. Novel viruses with high degree of overall similarity but featuring different RBP variants serve as useful model systems for in-depth analysis of T-even phage strategies used to circumvent O antigen protection of the host cell, efficiently shielded otherwise.
Three previously characterized E. coli strains featuring different O antigen types were used for bacteriophage isolation. These were: O28 strain F5 (Golomidova et al. 2019), E. coli F17, producing a novel O antigen type (Knirel et al. 2019) and O22 strain 4s (Knirel et al. 2015). To prepare concentrated phage stocks the the relevant host culture was propagated at 37°C with agitation up to OD600 = 0.2, and inoculated with phage at multiplicity of infection (moi) = 0.1. The culture was incubated under the same conditions until the lysis was observed, then 0.05 volume of chlorophorm was added and the lysates were cleared by centrifugation at 10 000 g for 10 min.
The bacteriophages were isolated by direct plating of the commercial therapeutical bacteriophage preparation “Intesti-bakteriofag” (Mikrogen, Russia) that is marketed as a broad range therapeutic phage cocktail (active against Shigella flexneri, S. sonnei, S. paratyphi A, S. paratyphi B, Salmonella typhimurium, S. infantis, S. choleraesuis, S. oranienburg, S. enteritidis, E. coli, P. vulgaris, P. mirabilis, Enterococcus sp., Staphylococcus, P. aeruginosa) (https://www.microgen.ru/products/bakteriofagi/intesti-bakteriofag/). This preparation was titrated to single plaques by the conventional double-layer plating technique using three above-mentiones E. coli strains as the hosts. All the methods for phage isolation were as described in Kulikova et al. (2007). PCR assay with RB49 g38 specific primers cttgctggatgagccaattcgccggacgctctaaagagattcattatagca-gp38C(RB49)-F and ccttttaagggtatttattcacattacacgcgagcaccgtaga-gp38C(RB49)-R was used to pick RB49-like phage candidates for further study.
Transmission electron microscopy study was performed using uranyl acetate negative contrast as described in (Kulikov et al. 2014).
To extract the DNA the lysates were treated with DNAse (0,01 mg mL−1) for 30 min at room temperature and the phages were collected by ultracentrifugation (1 h, 75000 g). Viral genomic DNA was extracted from the pellets with CTAB (cetyltrimethylammonium bromide) as described in (Kulikov et al. 2020) using. DNA quality and quantity was assessed by agarose gel analysis and Qubit dsDNA HS fluorometer assay (Qubit, USA). The phage genomic DNA libraries were prepared and sequenced using an Ion Torrent sequencer system (Applied Biosystems, USA) with 400-fold coverage and a median read length of 185 bp. The raw reads from the run were then combined and filtered using the spectral alignment error correction tool SAET 3 (Pevzner et al. 2001). This yielded 157,407-163,568 reads per library with average 190-fold coverage. Primary assembly was conducted with Newbler version 3.0 (Roche Diagnostics, USA), resulting in a single contigs.
Annotation was performed using Prokka (Seemann 2014) with further manual curation. Potential open reading frames (ORFs) were detected by use of SnapGene (GSL Biotech, USA) and subsequently analyzed with HMMER, HHPRED (Soding et al. 2005) (MPI Bioinformatics Toolkit), BLAST (Altschul et al. 1990), tRNAscan-SE (Lowe and Eddy 1997).
A subset of 25 highly similar RB49-like phages using with blastN search with RB49 genome sequence as a query was retrieved from GenBank. The thresholds used were 93% of the query coverage and the average nucleotide identity over aligned segments 92%. Full genome nucleotide alignment and phylogenetic tree reconstruction were performed with NGPhylogeny.fr web services. The genome of RB49 and the bacteriophage genomes obtained in this work were aligned and visualized with Clinker v0.0.21 and clustermap.js (Gilchrist and Chooi 2021).
The direct plating of the commercial therapeutical phage mixture on the lawns of the E. coli strains 4s, F5 and F17 yielded phage plaques. PCR screening with the primers hybridizing to bacteriophage RB49 g38 sequence was performed and about half of the plaques were found positive. One PCR-positive phage isolate per host strain was selected and purified by repeated single plaque isolation. The obtained bacteriophage strains were named as follows: F5-derived phage – Cognac49, F17-derived phage – Whisky49 and 4s-derived phage – Brandy49. The concentrated phage stocks were grown and analyzed by transmission electron microscopy. The bacteriophages were morphologically undistinguishable (Figure 1) having typical appearance of T-even-like viruses (Ackermann and Krisch 1997, Comeau et al. 2007) that was compatible with their expected relatedness to the phage RB49.
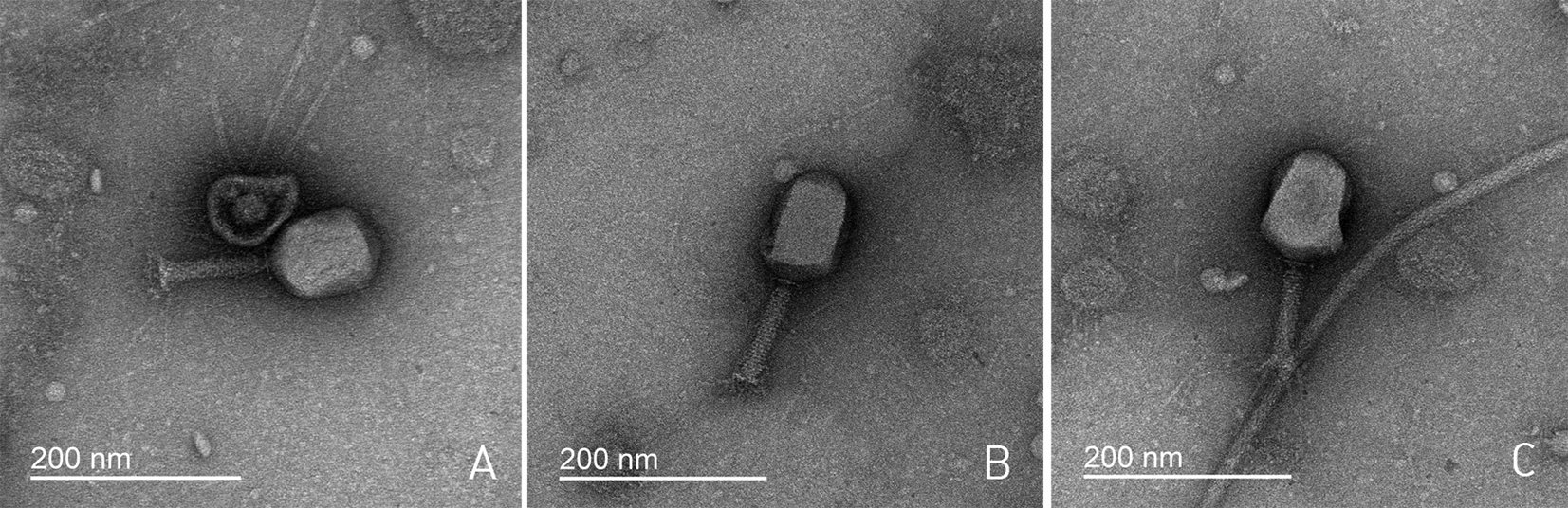
The genomic DNA was extracted from the phage stocks and sequenced using the Ion Torrent technology. The assembly procedure yielded a single contig for each of the bacteriophages with the coverage level > 190. Since T-even related phages feature unlimited circular permutations (Comeau et al. 2007), the start of each genomes was set arbitrary according to the standard representation of the RB49 genome. The main features of the genomes assembled are listed in the Table 1.
| Phage | Genome length, bp | Number of ORFs annotated | Average GC content, % | GenBank accession number |
|---|---|---|---|---|
| Brandy49 | 169,233 | 279 | 41 | MZ504876 |
| Cognac49 | 164,033 | 271 | 40 | MZ504877 |
| Whisky49 | 165,617 | 278 | 40 | MZ504878 |
Gene functions were predicted based on the existing annotations of RB49-like phages producing the best blastN hits for each of the genomes for Brandy49: Phi1 (nucleotide identity 95.77%), vB_EcoM_011D4 (95.57%), kaaroe (96.86%); for Cognac49: Sf20 (98.04%), RB49 (97.59%), KFS-EC (97.79%); for Whisky49: vB_EcoM_011D4 (97.76%), Sf20 (98.38%), GEC-3S (98.17%); query cover >95% for all comparisons. The annotations were then manually curated.
Scan by tRNAscan-SE detected no tRNAs in phage genomes.
The comparison of three genomes to the bacteriophage RB49 (Figure 2) revealed that 42 ORFs shared identical aa sequences in all four genomes, another 140 ORFs were also very similar with the aa identity ≥ 0.90. The phages share also 71 more distantly related ORFs with aa identity levels ≥ 0,50 but ≤ 0.90. This result indicates that the isolated bacteriophages are closely related to each other.
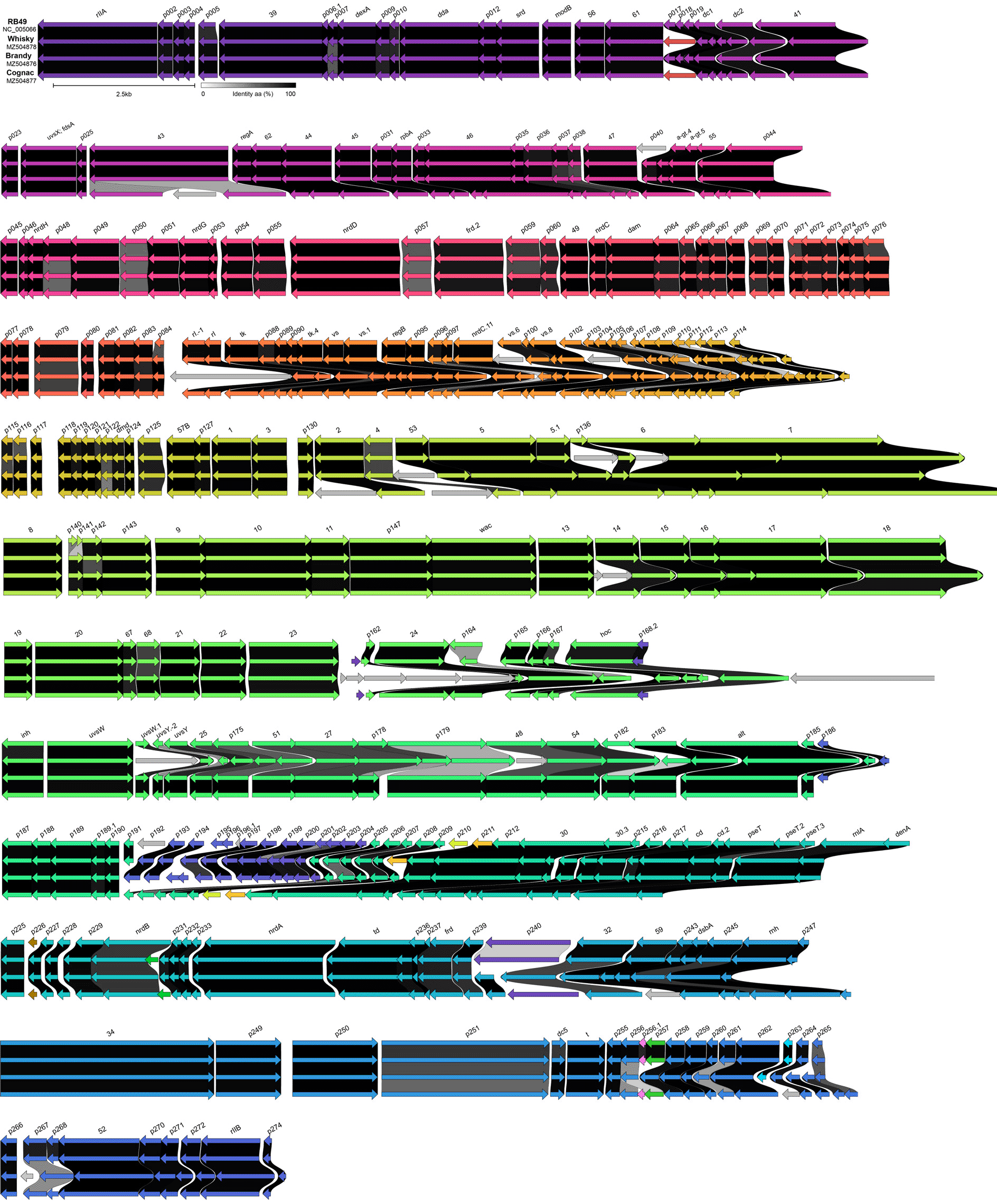
The grayscale indicates the level of the amino acid identity. The ORF colors reflect the groups of homology as revealed by Clinker software.
About 56 ORFs making the difference between these three phages (Figure 2), mostly encode for hypothetical proteins with unknown function. Not all RB49 ORFs or their homologues are present in all phages. A trace of recombination event was detected in genome of Cognac49 phage. An integration of foreign genetic material occurred within DNA polymerase gene, which is essential for phage viability. This insertion contains an 825 bp ORF predicted to be an HNH homing nuclease, often present within self-splicing introns. Rfam search (Kalvari et al. 2021) yielded no significant hits, while the analysis of putative secondary structures of corresponding RNA revealed a complex pattern of the stem-loop structures suggesting that insertion could be a bacterial type I intron containing homing nuclease. Ten ORFs with unknown function present in RB49, Brandy49 and Whisky49 are missing in genome of Cognac49, spanning the region homologous to NP_891764.1-NP_891775.1 in RB49.
High similarity of the studied phages to RB49 allows us to classify them as follows: Realm: Duplodnaviria, Kingdom: Heunggongvirae, Phylum: Uroviricota, Class: Caudoviricetes, Order: Caudovirales, Family: Myoviridae, Subfamily: Tevenvirinae, Genus: Krischvirus; Escherichia virus RB49 type viruses, just as expected from g38 PCR and TEM results. The genomes of 25 bacteriophages closely related to RB49 were extracted from GenBank. Overall nucleotide phylogeny tree of this subset of the phage genomes with the addition of three genomes sequenced during this work is shown on the Figure 3.
Despite their close relatedness, the isolated bacteriophages showed marked differences in their host specificity. When they were cross-tested for the infectivity on all three host strains used, all three phages were found infective towards F5 strain, however, phage Whisky49 could form plaque on both E. coli F5 and F17 strains, while Brandy49 had the broadest host range infecting all three host strains (been the only one forming plaques on the E. coli 4s lawns). In all the cases the efficiency of plating was similar that observed on the respective isolation host strain.
The specificity of host cell recognition by most of the T-even-like phages is determined by two proteins – the LTF adhesin gp38 and by the short tail fiber protein gp12 (PMID: 21746838). The comparison of gp12 aa sequences from the bacteriophages under the study revealed them to be nearly identical to gp12 from RB49 phage (data not shown). At the same time the sequences of gp38 were found more diverged (Figure 4). Gp38 from the phage Brandy49 was more distant from the consensus sequence compared to other two viruses (Figure 4) that is correlated with a broader host range of this phage. Some of the aa polymorphisms were observed within the loops located between polyglycine stretches and predicted to be responsible for interactions with the receptor (Trojet et al. 2011, Dunne et al. 2018). Therefore, differences in the particular regions of gp38 sequence may explain different host specificities of these bacteriophages.
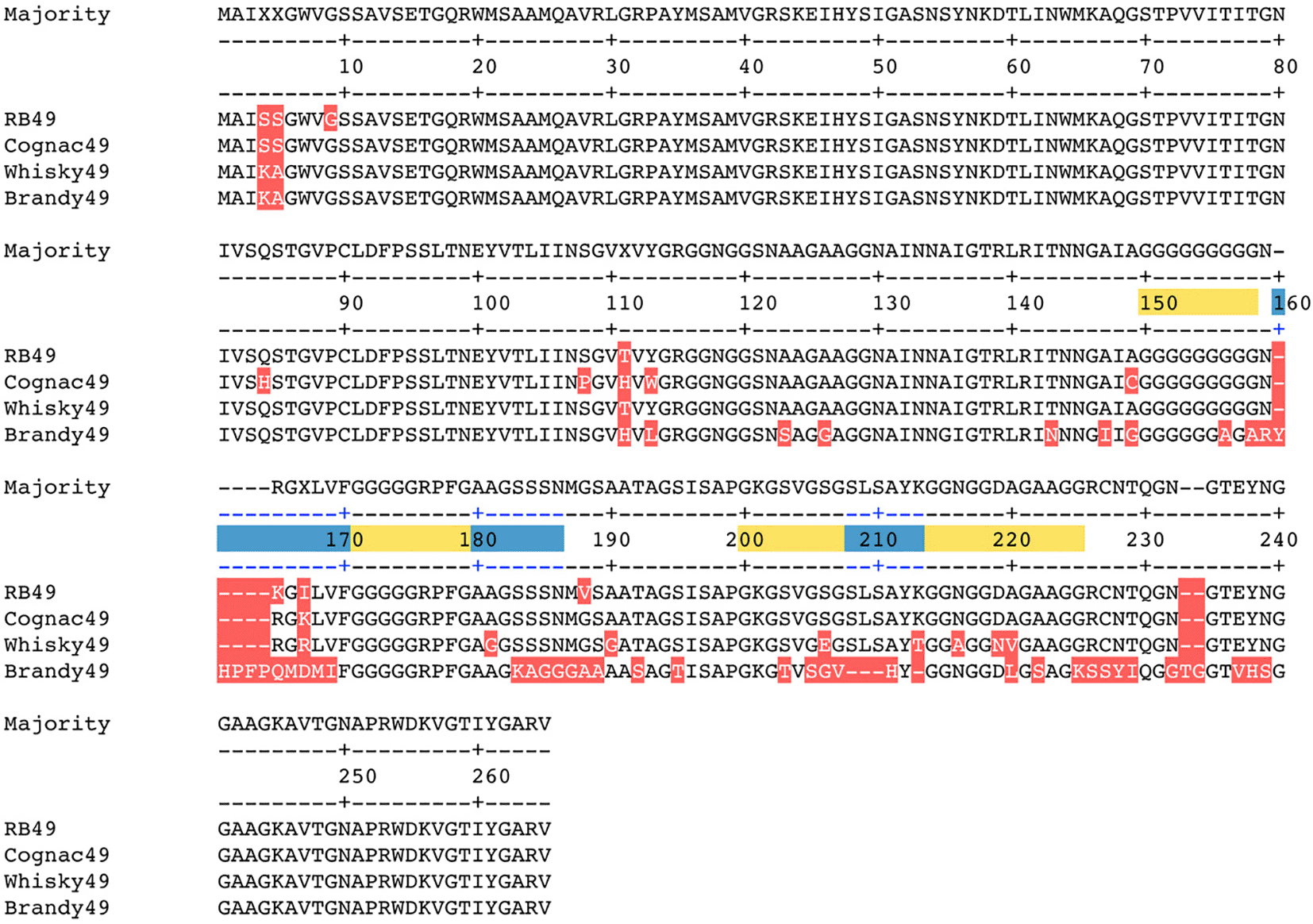
The aa residues that differ from the consensus are highlighted in red. Blue bars indicate polyglycine stretches and yellow bars the loops according to the structural data on phage S16 gp38 (Dunne et al. 2018).
The characterization of three new viruses that been closely related to each other show distinct patterns of the infectivity towards E. coli strain producing different types of the O antigen provides a useful model system for in-depth analysis of the strategies employed by T-even like phages to infect the host cells featuring effective non-specific protection of the outer membrane surfaces by this polysaccharide structure. The close relatedness of these phages to each other will facilitate the comparison of the results of experiments aiming at deciphering fine mechanisms employed by RB49-like phages to penetrate the O antigen shields of different E. coli strains.
Repository: NCBI Nucleotide database
Accession number: MZ504876 for Brandy49
Root URL: https://www.ncbi.nlm.nih.gov/nuccore
Accession number URL: https://www.ncbi.nlm.nih.gov/nuccore/MZ504876
Accession number: MZ504877 for Cognac49
Root URL: https://www.ncbi.nlm.nih.gov/nuccore
Accession number URL: https://www.ncbi.nlm.nih.gov/nuccore/MZ504877
Accession number: MZ504878 for Whisky49
Root URL: https://www.ncbi.nlm.nih.gov/nuccore
Accession number URL: https://www.ncbi.nlm.nih.gov/nuccore/MZ504878
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
References
1. Salem M, Pajunen M, Jun J, Skurnik M: T4-like Bacteriophages Isolated from Pig Stools Infect Yersinia pseudotuberculosis and Yersinia pestis Using LPS and OmpF as Receptors. Viruses. 2021; 13 (2). Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular microbiology, bacterial genetics, bacteriophages, phage therapy
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bacteriophage biology, phage-bacterial host interaction, prophage induction
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 03 Nov 21 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)