Keywords
protein-ligand binding, residence time, binding kinetics, web services, database
This article is included in the Cheminformatics gateway.
This article is included in the Bioinformatics gateway.
protein-ligand binding, residence time, binding kinetics, web services, database
The changes made include the addition of new references for a more critical approach to the residence time model.
See the authors' detailed response to the review by Wiesław Nowak
Current knowledge allowing for the determination of the effectiveness of small molecules in vivo is limited. It is known that the leading reason for drug candidate failure is the lack of efficacy caused by a poor translation of in vitro potency assays into in vivo activity in humans (Copeland, 2016a, 2016b; Swinney, 2009). In vitro experimentation refers to closed system conditions in which the drug molecule and its target are present at unchanging concentrations throughout the experiment (Tummino & Copeland, 2008). However, in living organisms, processes run under open, non-equilibrium conditions where the drug is constantly interacting with various molecules during many physiological processes in addition to its native target. To improve prediction of in vivo drug efficacy measurements of drug-target complexes, the residence time, defined as the reciprocal of the dissociation rate constant (koff), should be considered. Drug-target residence time is crucial because pharmacological activity depends on the drug being bound to its molecular target. When the drug dissociates from the binding site, the target molecule is free to continue its pathophysiological function. Within 10 years of the development of the drug-target residence time concept, the parameter has become an extremely important factor in the process of optimizing lead structures in computer-aided drug design (Copeland et al., 2006; Copeland, 2016a, 2016b). Traditional computational approaches (such as molecular docking) take into account only the equilibrium affinity of the drug for its molecular target (e.g., Ki); however, the concept of residence time also takes into account conformational dynamics of molecules, which can have a significant effect on the binding and dissociation of the drug (Copeland et al., 2006; Tummino & Copeland, 2008; Copeland, 2021). Nevertheless, there is some criticism indicating that using the residence time as the only measure of drug efficacy provides a limited picture of binding and affinity kinetics and is a suboptimal way to guide drug discovery programs (Folmer, 2018). Existing free and commercial software does not include a residence time model that is able to estimate this quantity, and thus the effects of ligand modification relevant to the design and optimization of drug candidates is not available.
To enable further studies and the development of useful drug-target residence time models, we created a database of experimentally measured residence times for biomolecular complexes deposited in the Protein Data Bank (PDB). These data represent a link between structural and kinetic information of the complexes which may be helpful for various computational and machine learning studies on drug-like molecules in biological systems. The current version of our database, called PDBrt, contains 59 complexes with experimentally measured residence time including seven protein families, and 56 small molecules. Summary statistics are available in Figure 1 and Table 1.
PDBrt database design and structure
The PDBrt database has been designed as an interactive web interface where the user can browse and extract information about the ligand residence time in its molecular target. The RESTful application programming interface (API) with the Django Rest Framework (v2.2.20) and a backend PostgreSQL (v12) database running on a Nginx (v1.21.3) web server has been developed to represent the output of a query as a user-friendly web page generated in Bootstrap (v3.3.7) with Hypertext Markup Language (HTML), Cascading Style Sheets (CSS) and jQuery (v3.5.1) to report the results. Search, query, and data extraction and visualization systems were developed for searching ligand residence time and binding kinetics coefficients. The PDBrt database facilitates access to information about the ligand residence time in its molecular target.
PDBrt database management system
The Database Management System (DBMS) allows users and programmers to manipulate the data in a systematic way. The DBMS serves as an interface between the database and end users, ensuring consistent data organization and easy accessibility. The PDBrt database is a type of relational DBMS using Structured Query Language (SQL) as the standard programming language for data manipulation.
Development of the PDBrt database was a multi-step process consisting of a systematic literature search, abstract and report screening, and article review. Several sequential steps involved in data management ensure that the processed data is accessible, reliable, and current for its users (Figure 2).
Binding kinetics data acquisition and extraction
Data acquisition starts with collection of protein-ligand binding kinetics data from available literature by reviewing the primary reference of each pdb file in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) to retrieve the experimentally measured ligand-target residence time or dissociation rate (koff). The references were downloaded and carefully studied to manually extract the data. Only complexes with known ligand-target residence time or dissociation rate were added into the final dataset. Additionally, major binding kinetics coefficients were collected (if available): inhibition constant (Ki) and association rate (kon). Currently the PDBrt database includes 59 protein-ligand complexes with known ligand-target residence time. Structures will be added on a regular basis as respective data becomes available.
Structural data acquisition
Complexes with particular PDB identifiers are downloaded from the RCSB PDB database into the internal PDBrt database core. The protein molecule along with other components such as water molecules and metal ions were saved in the pdb format, while the ligand (drug) in Structure Data Format (sdf). Neither the protein nor the ligand was subjected to any structural optimization or modification after being downloaded from the RCSB PDB.
PDBrt data content
PDBrt database data contain, in addition to the coordinates and general information required for all deposited structures in the RCSB PDB database, target residence time and other binding kinetics coefficients, structure files mentioned above, basic ligand properties like simplified molecular-input line-entry system (SMILES) or International Chemical Identifier (InChI) string, as well as links to external databases: RCSB PDB, PDBj, PDBe, PDBsum, and the reference literature in PubMed. The database includes citations to the original sources (publications) that contain information about the experimentally measured residence time or dissociation rate. For each complex, a web-based three-dimensional rendering is provided using the free, openly available object-oriented JavaScript library 3Dmol.js, which is used for visualizing molecular data.
PDBrt database availability and updates
PDBrt is available to the community through its web-based interface. Since the PDB database is growing rapidly and drug–target residence time is a parameter of great interest for drug design and optimization, the PDBrt will be updated on a regular basis with major versions issued annually. New data can be added by the authors (database administrators) either by uploading an xlsx (MS Excel) file or manually.
PDBrt database architecture
PDBrt is a three-tier architecture:
1. Data tier comprises database and data access layer.
2. Application tier controls application functionality by performing detailed processing.
3. Presentation tier is accessible for end-users and displays information on the website.
Core relational database managed by PostgreSQL server provides information storage for the deposited data. Back end (data access layer) was implemented in Python (v3.6) and the front end of the database (presentation layer) in HTML/CSS.
PDBrt database model
In PDBrt data is presented as a collection of relations - tables. Each column (also called attribute or field) in the table has a distinct name and a specific data type assigned to it. All the information related to a particular type is stored in a row (also called record) of that table. PDBrt database has three main columns (‘Complex’, ‘Protein’, ‘Ligand’) and 59 records with one-to-one type of relationship. This means that one protein or ligand could only belong to one complex and one complex consists of only one specific protein and ligand molecule (Figure 3).
PDBrt is available to the community through its web-based interface and is freely available to non-commercial users. The PDBrt database runs on all modern web browsers. See Software availability (Ługowska & Pacholczyk, 2021) for access to the database and code.
Use case
To show the usage of the PDBrt database, the Unified Modelling Language (UML) use case diagram was adopted. Figure 4 shows five use cases of PDBrt as well as three actors: system administrator, end user and the database. The database is an actor for all use cases, system administrator for two and end user for three of these. The database actor is involved in all five use cases because it stores the data and enables operations by which advanced data handling functions are created. In the ‘complex management’ use case the system administrator can perform basic Create, Read, Update and Delete (CRUD) actions on the ‘Complex’ table: add, delete, update as well as upload new data as Microsoft Excel (xlsx) file format. In the ‘user management’ use case the system administrator can add, edit, and delete a regular user. In the ‘view’, ‘search’ and ‘download’ use case a user actor is involved. In the ‘view’ use case a list of complexes is displayed to the user who can choose a single complex and view its details as well as its 3D visualization. In the ‘search’ use case the user can filter from the whole database by PDB code, residence time, and protein or ligand name. The ‘download’ use case allows users to either download the data stored in the database as plain text file or pdb/sdf file formats.
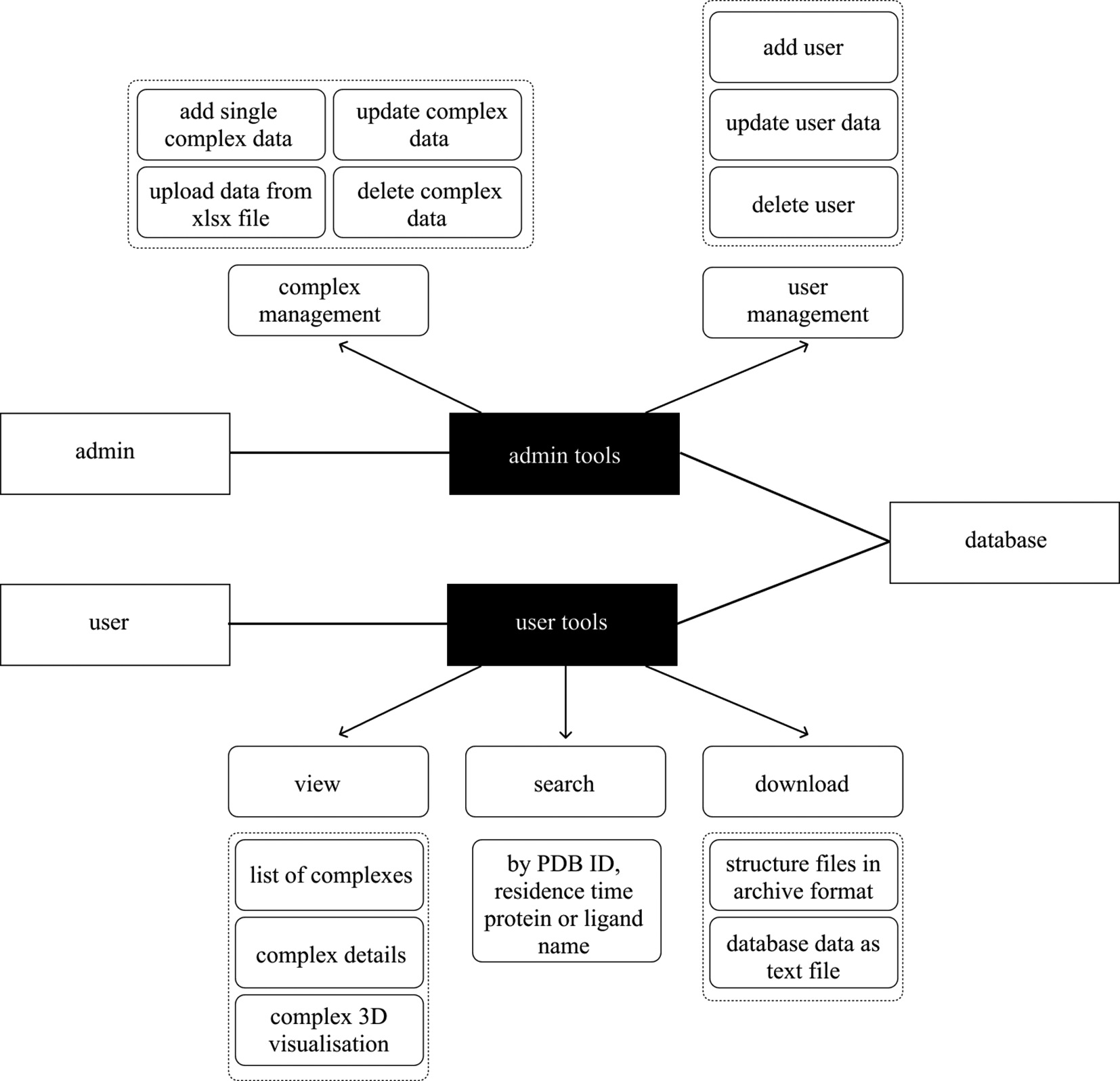
Diagram shows a subset of functions available to the regular user and website administrator.
‘Rectangle’ represents a user, ‘rounded rectangle’ represents a use case and ‘arrow’ represents a relationship.
Query interface has been implemented for the query of data within PDBrt. Figure 5 shows how the query options are organized. The search engine provides one form field for keyword search and allows retrieval by PDB code, protein name and residence time.
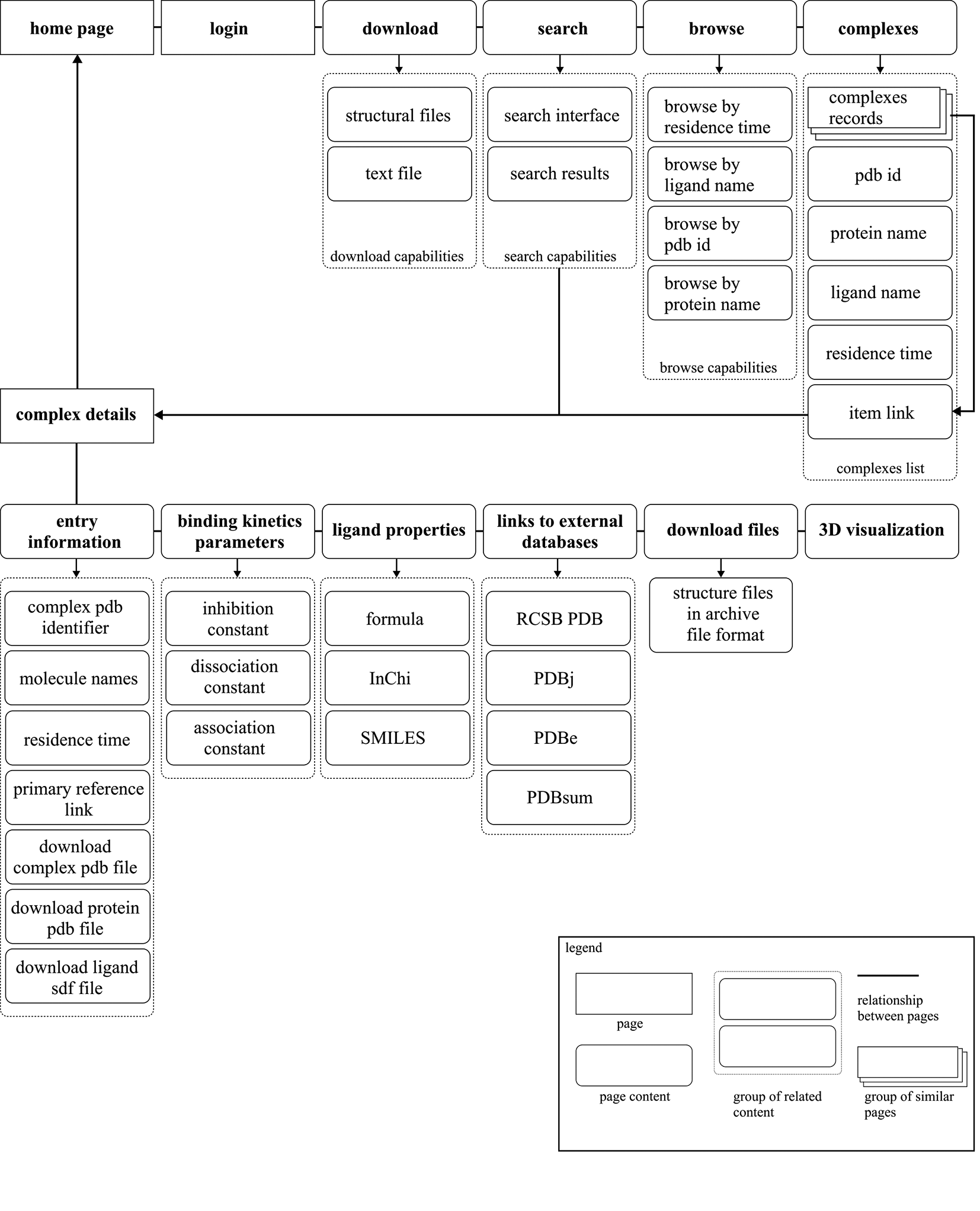
Diagram presents the structure of the PDBrt website. Each symbol (shape or arrow) presents PDBrt web page (single web view), page content, group of related content on a single page, relationship between web pages and group of similar web pages as described in the legend.
Two user interfaces provide extensive information for result sets obtained for particular search query. The ‘homepage’ interface allows access to some general information in tabular format and offers the possibility to download whole sets of data files for result sets consisting of multiple PDBrt entries. The ‘complex detail’ interface provides information about individual structures as well as cross-links to many external resources for macromolecular structure data.
Drug-target residence time has been shown to play an important role in the prediction of in vivo efficacy of the drug. Since the parameter is independent of drug and enzyme concentration, it is important in the drug design process and should be considered at an early stage. The availability of information about drug-target complexes with known (measured) residence times is becoming more and more significant. Several databases containing information about protein – ligand complexes are available, for example PDB-bind (Liu et al., 2015), BindingDB (Gilson et al., 2016), and Binding MOAD (Smith et al., 2019). However, these databases (except for BindingDB) do not store detailed information about binding rates, and none of these contain direct information about ligand (drug) residence time in its target macromolecule. The PDBrt database is dedicated to reporting the structure of protein - small molecule complexes, along with their target residence time and additional binding parameters (Ki, kon, koff). Currently a total of 59 protein–ligand complexes are deposited in the web – based PDBrt database and this data will be updated frequently as more data is made available. The information associated with the existing 59 protein–ligand complexes is available in Underlying data (Ługowska, 2021).
Zenodo: Drug-target residence time data. https://doi.org/10.5281/zenodo.5647983 (Ługowska, 2021).
This project contains the following underlying data:
‐ residence_time_data.xlsx (information about target residence time and other binding kinetics coefficients, basic ligand properties like simplified molecular-input line-entry system (SMILES) or International Chemical Identifier (InChI) string, as well as the reference literature in PubMed).
‐ structures.zip (the protein molecule along with other components such as water molecules and metal ions in the pdb format, the protein-ligand structure in the pdb format, and the ligand (drug) in both pdb and Structure Data Format (sdf)).
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Database available from: https://pdbrt.polsl.pl/
Source code available from: https://github.com/mlugowska/residence_time
Archived source code at time of publication: https://doi.org/10.5281/zenodo.5583543 (Ługowska & Pacholczyk, 2021)
License: MIT
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
References
1. Lu H, Iuliano JN, Tonge PJ: Structure-kinetic relationships that control the residence time of drug-target complexes: insights from molecular structure and dynamics.Curr Opin Chem Biol. 2018; 44: 101-109 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: I am structural bioinformatician, but do not have much experience with kinetic measurements
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: biophysics, bioinformatics, computer modeling of proteins, single molecule nanomechanics, atomic and molecular physics
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Partly
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
References
1. Śledź P, Caflisch A: Protein structure-based drug design: from docking to molecular dynamics.Curr Opin Struct Biol. 48: 93-102 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: biophysics, bioinformatics, computer modeling of proteins, single molecule nanomechanics, atomic and molecular physics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 31 Oct 22 |
read | read |
|
Version 1 03 Dec 21 |
read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)