Keywords
ABCG2, SLC22A12, XDH, hyperuricemia, hypercholesterolemia
This article is included in the Cell & Molecular Biology gateway.
ABCG2, SLC22A12, XDH, hyperuricemia, hypercholesterolemia
This new version shows minor changes and clarifications noted by the reviewer 2. The number of cases and controls as well as the description of sequence variants were included in the abstract. Some issues related to the statistical power and the genotype-based mRNA expression analysis were discussed in greater depth.
See the authors' detailed response to the review by Ilham Akbar Rahman
See the authors' detailed response to the review by Jing He
Hyperuricemia is an abnormal metabolic trait defined as serum uric acid levels above 6 and 7 mg/dL for women and men, respectively (Bardin & Richette, 2014), and is associated with other cardiometabolic risk factors such as obesity, diabetes, hypertension, and dyslipidemia (Martínez-Quintana et al., 2016; Zuo et al., 2016). Similarly, hyperuricemia can be a predictor of impaired kidney functions as well as kidney disease progression (Galán et al., 2018; Hsu et al., 2009, and Pérez-Navarro et al., 2020). Serum uric acid levels depend on many factors including diet, sex, lifestyle, and alcohol consumption, as well as genetic heredity. In fact, some genes involved in the metabolism of purines or urates have versions that appear to predispose to a hyperuricemic condition. Some of these are the ABCG2 gene that encodes a membrane transporter, which exports urates to the kidney and intestine (Woodward et al., 2009); and the SLC2A9 gene that encodes a kidney protein called GLUcose Transporter 9 (GLUT9), an important flow regulator of urates in the proximal tubules (Caulfield et al., 2008). In the same way, the XDH gene gives rise to an enzyme denominated xanthine dehydrogenase, which breaks down purines from nucleic acids, specifically the hypoxanthine‒xanthine‒urate conversion pathway (Harrison, 2002).
Recent studies have shown that genetic polymorphisms in ABCG2 and SLC2A9 genes are prevalent in the Mexican population and may contribute to an abnormal condition of hyperuricemia, even at young ages (Macías-Kauffer et al., 2019; Rivera-Paredez et al., 2019). Interestingly, there appears to be no reports to date in the literature on XDH gene polymorphisms in the Mexican population. Therefore, the aim of this study was to genotype the polymorphisms of the genes ABCG2 (Q191K), SLC22A12 (517G>A), and XDH (518T>C) in 860 young Mexican volunteers aged between 18 and 25 years of age as predisposing factors of hyperuricemia associated with risk factors of cardiovascular disease.
A cross-sectional design and a convenience sample of 860 subjects was coducted in the City of San Luis Potosí, México. All participants were college applicants during the spring period of 2017. Data collection was carried out according to the recruitment protocols of the University of San Luis Potosí, since applicants receive a clinical evaluation as part of the admission process. This project was approved by two Bioethics Committees: the former at the National Institute of Medical Sciences and Nutrition Salvador Zubirán (Approval # FNU-669-13-15-2), and the latter at the Faculty of Chemistry of the University of San Luis Potosí (Approval # CEID2017105-S). All participants provided written informed consent.
A total of 860 applicants to the state University of San Luis Potosí, Mexico in 2017, were included in this study (448 men and 412 women). All participants were approached to take part in the study by phone and email by means of an open invitation explaining the goals of the study. Even though the invitation was open, only some accepted to participate. Participants were not paid to take part. All individuals who provided written informed consent and met the inclusion criteria were included. The inclusion criteria which were as follows: aged between 18–25 years; born in the Mexican state of San Luis Potosí; and provided written informed consent.
Since all participants at the University of San Luis Potosí, Mexico receive a clinical evaluation as part of the admission process, we accessed the university medical records. The weight in kilograms and height in meters of each participant was obtained for anthropometry using a digital scale (UM-081 model; Tanita, Tokyo, Japan) and a stadiometer (Seca 213, 2009; Seca, Hanover, MD, USA), respectively. From this data, the Body Mass Index (BMI) was calculated (kg/m2). Systolic and diastolic blood pressure were taken as the mean of two readings at a 5-min interval after 5 min in a seated position, employing the Omrom model HBP-1300 portable meter (Omron Healthcare, Inc., IL, USA). Also, 6 mL of blood was obtained by venipuncture after a 12-h fast; serum was obtained by centrifuging at 1,000 x g for 10 min using a laboratory centrifuge Z306 Benchmark Scientific (NY, USA), and processed inmmediately. An addition, 3 mL of blood was collected and stored in ethylenediaminetetraacetic acid (EDTA) tubes (Vacutainer®) for subsequent DNA purification. All samples were maintained at -20°C until their analysis.
Serum was used to measure uric acid, glucose, total cholesterol, low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), and triglycerides by spectrophotometry utilizing chemistry reagents (Paramedical S.r.l., Italy), specially developed to work with Mindray BS 300 Auto Chemistry Analyzer (Mindray, Shenzhen, China), with the following catalog numbers: uric acid (PDIBS200040), glucose (PDIBS200020), total cholesterol (PDIBS200030), LDL-C (PDIBS200170) HDL-C (PDIBS200160), and triglycerides (PDIBS200060).
DNA was isolated from whole blood using the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany). The quality of DNA samples was verified by spectrophotometry using NanoDrop™ 2000. Polymorphisms of the genes ABCG2 (rs2231142), SLC22A12 (rs476037), and XDH (rs1042039) were measured by allelic discrimination real-time polymerase chain reaction (PCR) with the Taqman probe (Applied Biosystems®).
All PCR reactions were carried out using an ABI Prism 7900 HT detection system in 96-well plates (Applied Biosystems®). Reactions were adjusted to a final volume of 10 µL including Master mix probes and 50 ng/µL of genomic DNA, with the following amplification protocol: denaturation 94° for 9 min; followed by 50 cycles of denaturation at 95°C for 30 s, and annealing and extension at 68°C for 11 min. Negative controls and duplicated samples were included to check the accuracy of the genotyping.
After analyzing the data distribution, comparison of medians between groups (normal uricemia vs. hyperuricemia) was carried out with the Mann‒Whitney U test for continuous variables. Allelic and genotypic frequencies as well as the Hardy‒Weinberg equilibrium were calculated for each polymorphism utilizing the chi-square test. Likewise, a logistic regression analysis was used to calculate the risk of hyperuricemia in relation to the variables studied and the influence of the different genotypes on uric acid levels in blood through a dominant model. The level of significance was set at p <0.05. Data were analyzed using SPSS version 19.0 statistical software.
In total, 860 participants were included in this study: 52% men and 48% women (Alegría-Torres, 2021). Participant characteristics, as well as the allelic and genotypic frequencies of the ABCG2 (Q191K), SLC22A12 (517G>A) and XDH (518T>C) gene polymorphisms, are presented in Table 1. Medians and genotypic distribution between hyperuricemic and normouricemic participants were analyzed using the Mann-Whitney U test, finding differences for sex, BMI, systolic and diastolic pressure, triglycerides, total cholesterol, and LDL-C (p = <0.001 for all), as well as a marginal distribution difference for the ABCG2 (Q191K) polymorphism corresponding to G/T alleles (p = 0.056), therefore other genetic models were tested later. All the genotypes analyzed were in Hardy‒Weinberg equilibrium: ABCG2 (Q191K) X2 = 1.82X10-6, p = 0.9; SLC22A12 (517G>A) X2 = 0.03, p = 0.84; and XDH (518T>C) X2 = 0.08, p = 0.76. Both allelic and genotypic distributions of the three polymorphisms studied did not differ significantly between men and women (Table 2).
| Variable | All participants median (p25-p75) n (%) | With hyperuricemia median (p25-p75) n(%) | With normal uricemia median (p25-p75) n(%) | P value |
|---|---|---|---|---|
| Total Men Women | 860 (100) 448 (52) 412 (48) | 129 (15) 88 (19.6) 41 (10) | 731 (85) 360 (80.4) 371 (90) | <0.001a |
| Age (years) | 19.0 (18.0-20.0) | 19.0 (18.0-20.0) | 19.0 (18.0-20.0) | 0.138b |
| BMI (Kg/m2) | 23.03 (20.56-26.58) | 25.7 (23.0-29.7) | 22.6 (20.2-26.1) | <0.001b |
| Systolic pressure (mmHg) | 110 (100-110) | 110 (100-120) | 110 (100-110) | <0.001b |
| Diastolic pressure (mmHg) | 70 (60-70) | 70(70-80) | 70 (60-70) | <0.001b |
| Glucose (mg/dL) | 79.0 (74.5-84.0) | 81.0 (75.0-86.0) | 79.0 (74.0-84.0) | 0.087b |
| Uric acid (mg/dL) | 5.3 (4.2-6.2) | 7.6 (6.9-8.2) | 4.9 (4.1-5.7) | <0.001b |
| Triglycerides (mg/dL) | 96.0 (71.0-133.0) | 126.0 (96.0-178.0) | 90.0 (68.0-126.5) | <0.001b |
| Total Cholesterol (mg/dL) | 150.0 (131.0-170.0) | 164.0 (144.0-188.5) | 146.0 (128.0-166.0) | <0.001b |
| LDL Cholesterol (mg/dL) | 58.5 (40.8-70.05) | 69.1 (49.7-94.3) | 57.1 (40.2-73.0) | <0.001b |
| HDL Cholesterol (mg/dL) | 67.9 (58.8-76.9) | 64.8 (56.1-74.9) | 68.3 (59.3-76.9) | 0.083b |
| ABCG2 (Q191K) Alleles G T Genotypes GG GT TT | 1298 (75) 422 (25) 490 (57) 318 (37) 52 (6) | 183 (71) 75 (29) 64 (49.6) 55 (42.5) 10 (7.9) | 1115 (76) 347 (24) 426 (58) 263 (36) 42 (6) | 0.056a 0.17a |
| SLC22A12 (517G>A) Alleles G A Genotypes GG GA AA | 558 (32) 1162 (68) 94 (11) 370 (43) 396 (46) | 84 (33) 174 (67) 15 (11.8) 54 (41.7) 60 (46.5) | 474 (32) 988 (68) 79 (11) 316 (43) 336 (46) | 0.9a 0.94a |
| XDH (518T>C) Alleles T C Genotypes TT TC CC | 1083 (63) 637 (37) 335 (39) 413 (48) 112 (13) | 167 (65) 91 (35) 53 (41) 61 (47.2) 15 (11.8) | 916 (63) 546 (37) 282 (39) 352 (48) 97 (13) | 0.52a 0.8a |
Abbreviations: BMI, body mass index; ABCG2, gene that encodes an ATP-binding cassette transporter subfamily G member 2; SLC22A12, gene that encodes an urate transporter 1 gene; XDH, gene that encodes a xanthine dehydrogenase.
aChi-square test.
bMann‒Whitney U test.
cSerum uric acid ˃6 and 7 mg/dL was considered as hyperuricemia for women and men, respectively.
Data are shown as median and 25th and 75th percentile range (p25–p75).
The level of significance was set at p <0.05.
| Men n(%) | Women n (%) | P valuea | |
|---|---|---|---|
| ABCG2 (Q191K) Alleles G T Genotypes GG GT TT | 683 (76) 213 (24) 255 (57) 173 (39) 20 (4) | 615 (75) 209 (25) 233 (57) 149 (36) 30 (7) | 0.44 0.19 |
| SLC22A12 (517G>A) Alleles G A Genotypes GG GA AA | 286 (32) 610 (68) 47 (10) 192 (43) 209 (47) | 272 (33) 552 (67) 49 (12) 174 (42) 189 (46) | 0.63 0.80 |
| XDH (518T>C) Alleles T C Genotypes TT TC CC | 567 (63) 329 (37) 181 (40) 205 (46) 62 (14) | 516 (63) 308 (37) 156 (38) 204 (50) 52 (12) | 0.77 0.54 |
Considering the statistically significant differences among the variables, a multivariate logistic regression analysis was performed, including sex, BMI, hypercholesterolemia, and hypertriglyceridemia as predictors of hyperuricemia (Figure 1). A predisposition to abnormal serum uric acid levels was found in some conditions as follows: male sex (odds ratio (OR): 2.14, 95% confidence interval (CI): 1.41-3.24, p = ˂ 0.001); overweight or obese (OR: 2.6, 95% CI: 1.71-3.88, p = ˂0.001); having hypercholesterolemia (OR: 2.8, 95% CI: 1.47-5.46, p = 0.002); or having hypertriglyceridemia (OR: 1.7, 95% CI: 1.04-2.69, p = 0.033).
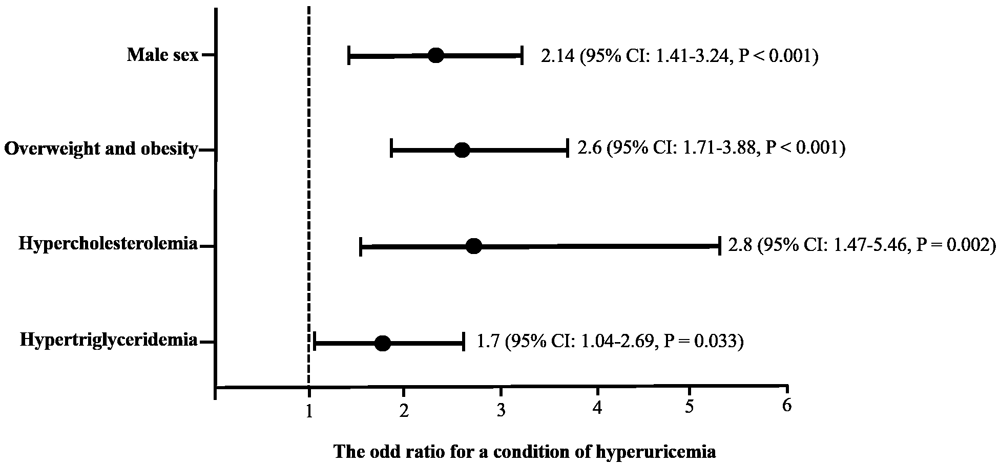
The cutoff criteria were based on the World Health Organization and the National Cholesterol Education Program Adult (ATP III). Overweight: body mass index ≥ 25, Obesity: body mass index ˃30; hypercholesterolemia: fasting serum cholesterol levels ˃ 200 mg/dL; hypertriglyceridemia: fasting serum triglycerides levels ˃ 150 mg/dL.
In Table 3, an analysis by genotype is shown for the ABCG2 (Q191K), SLC22A12 (r517G >A), and XDH (518T>C) polymorphisms, employing a dominant model for all three. With respect to the ABCG2 (Q191K) polymorphism, statistically significant differences were found between the GT+TT genotypes vs. GG for serum uric acid (p = 0.003) total cholesterol (p = 0.005) and HDL-C levels (p = 0.003). GA+AA genotypes vs. GG of the SLC22A12 (517G>A) polymorphism only showed to influence LDL-C levels (p = 0.043). Finally, no statistically significant differences were found by genotype for the XDH (518T>C) polymorphism (p > 0.05).
| Variable | ABCG2 (Q191K) Dominant model | P-Valuea | |
|---|---|---|---|
| GT+TT median (25th-75th percentile range) | GG median (25th-75th percentile range) | ||
| BMI (Kg/m2) | 23.1 (20.4 – 27.0) | 22.7 (20.2 – 26.3) | 0.393 |
| Systolic pressure (mmHg) | 110 (100.0 – 110.0) | 110 (100.0 – 110.0) | 0.632 |
| Diastolic pressure (mmHg) | 70.0 (60.0 – 70.0) | 70.0 (60.0 – 70.0) | 0.220 |
| Glucose (mg/dL) | 80.0 (76.0 – 85.0) | 80.0 (75.0 – 84.0) | 0.129 |
| Uric Acid (mg/dL) | 5.00 (4.2 – 6.2) | 4.7 (3.9 – 5.4) | 0.003 |
| Triglycerides (mg/dL) | 94.0 (69.5 – 134.0) | 92.0 (68.0 – 127.0) | 0.634 |
| Total Cholesterol (mg/dL) | 151 (132.0 – 173.0) | 145 (125.0 – 164.0) | 0.005 |
| LDL Cholesterol (mg/dL) | 59.2 (42.5 – 74.8) | 53.8 (38.8 – 72.7) | 0.070 |
| HDL Cholesterol (mg/dL) | 70.9 (62.3 – 78.5) | 67.3 (57.8 – 76.8) | 0.003 |
| SLC22A12 (517G>A) Dominant model | |||
| GA+AA median (p25-p75) | GG median (p25-p75) | ||
| BMI (Kg/m2) | 22.6 (20.1–26.2) | 23.2 (20.4–26.6) | 0.136 |
| Systolic pressure (mmHg) | 110.0 (100.0 – 110.0) | 110.0 (100.0 – 110.0) | 0.662 |
| Diastolic pressure (mmHg) | 70.0 (60.0 – 70.0) | 70.0 (60.0 – 70.0) | 0.600 |
| Glucose (mg/dL) | 80.0 (76.0 – 85.0) | 80.0 (75.0 – 84.5) | 0.302 |
| Uric Acid (mg/dL) | 4.8 (4.0 – 5.5) | 4.90 (4.1 – 5.6) | 0.537 |
| Triglycerides (mg/dL) | 91.0 (68.0 – 135.0) | 94.5 (70.0 – 125.5) | 0.763 |
| Total Cholesterol (mg/dL) | 144.0 (126.0 – 167.0) | 149.5 (131.5 – 169.0) | 0.127 |
| LDL Cholesterol (mg/dL) | 54.3 (36.1 – 70.5) | 58.4 (40.6 – 75.2) | 0.043 |
| HDL Cholesterol (mg/dL) | 69.3 (59.1 – 77.6) | 68.1 (59.8 – 77.1) | 0.659 |
| XDH (518T>C) Dominant model | |||
| TC+CC median (p25-p75) | TT median (p25-p75) | ||
| BMI (Kg/m2) | 22.8 (20.2–26.4) | 23.0 (20.4–26.6) | 0.618 |
| Systolic pressure (mmHg) | 110.0 (100.0 – 110.0) | 110.0 (100.0 – 110.0) | 0.460 |
| Diastolic pressure (mmHg) | 70.0 (60.0 – 70.0) | 70.0 (60.0 – 70.0) | 0.949 |
| Glucose (mg/dL) | 80.0 (75.0 – 85.0) | 79.5 (76.0 – 84.0) | 0.521 |
| Uric Acid (mg/dL) | 4.80 (4.0 – 5.5) | 4.90 (4.0 – 5.5) | 0.840 |
| Triglycerides (mg/dL) | 94 (69.0 – 128.5) | 91.0 (68.0 – 132.0) | 0.944 |
| Total Cholesterol (mg/dL) | 147.0 (128.0 – 168.0) | 145.5 (129.0 – 167.0) | 0.564 |
| LDL Cholesterol (mg/dL) | 56.6 (39.5 – 73.3) | 56.1 (39.1 – 72.9) | 0.640 |
| HDL Cholesterol (mg/dL) | 69.1 (60.1 – 77.6) | 67.4 (58.9 – 76.9) | 0.370 |
When the OR and the association between genotypes for the BCG2 (Q191K) and SLC22A12 (517G>A) polymorphisms and cardiovascular risks were calculated by sex, some significant results were found utilizing a dominant model. Table 4 reports that GT+TT genotypes for the ABCG2 (Q191K) polymorphism significantly statistically increases the risk of hyperuricemia (OR = 2.43, 95% CI: 1.41-4.17, p = 0.001) and hypercholesterolemia (OR = 4.89, 95% CI: 1.54-15.48, p = 0.003), but only in male participants. On the other hand, no significant results were found for the genotypes of the SLC22A12 (517G>A) and XDH (518T>C) polymorphisms.
| Conditiona | Dominant model for the ABCG2 (Q191K) polymorphism GT+TT vs. GG | |||||
|---|---|---|---|---|---|---|
| All participants | Men | Women | ||||
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |
| Overweight and obesity | 1.10 (0.78-1.56) | 0.559 | 1.57 (0.93-2.67) | 0.089 | 0.84 (0.53-1.34) | 0.481 |
| SP ˃130 mmHg | 1.15 (0.38-3.47) | 0.799 | 2.81 (0.68-11.52) | 0.135 | 0.32 (0.03-2.97) | 0.298 |
| DP ˃85 mmHg | 0.33 (0.03-2.99) | 0.303 | 1.35 (0.08-21.94) | 0.829 | 0.44 (0.04-4.28) | 0.468 |
| Fasting glucose ˃100 mg/dL | 4.1 (0.82-20.49) | 0.063 | 2.74 (0.24-30.63) | 0.394 | 5.4 (0.60-49.39) | 0.091 |
| Hyperuricemia | 1.49 (1.00-2.21) | 0.047 | 2.43 (1.41-4.17) | 0.001 | 0.77 (0.39-1.53) | 0.465 |
| Hypertriglyceridemia | 0.84 (0.54-1.31) | 0.463 | 1.32 (0.76-2.30) | 0.315 | 0.87 (0.50-1.51) | 0.635 |
| Hypercholesterolemia | 3.27 (1.62-6.59) | 0.001 | 4.89 (1.54-15.48) | 0.003 | 2.09 (0.83-5.26) | 0.110 |
| LDL-C ≥130 mg/dL | 3.1 (0.81-12.47) | 0.078 | 5.59 (0.61-50.80) | 0.086 | 2.0 (0.33-12.26) | 0.435 |
| Low HDL-C | 0.37 (0.07-1.83) | 0.210 | 0.72 (0.319-1.644) | 0.440 | 0.44 (0.045-4.2) | 0.468 |
| Conditiona | Dominant model for the SLC22A12 (517G>A) polymorphism GA+AA vs. GG | |||||
| All participants | Men | Women | ||||
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |
| Overweight and obesity | 0.81 (0.57-1.14) | 0.232 | 0.72 (0.42-1.22) | 0.227 | 0.88 (0.56-1.39) | 0.599 |
| SP ˃130 mmHg | 1.01 (0.33-3.04) | 0.986 | 1.09 (0.28-4.19) | 0.892 | 0.85 (0.11-6.15) | 0.878 |
| DP ˃85 mmHg | 1.30 (0.21-7.84) | 0.773 | 1.76 (0.15-19.69) | 0.641 | 0.42 (0.03-4.74) | 0.475 |
| Fasting glucose ˃100 mg/dL | 2.6 (0.52-13.13) | 0.222 | 0.43 (0.03-4.84) | 0.485 | 4.3 (0.50-37.99) | 0.143 |
| Hyperuricemia | 0.99 (0.67-1.48) | 0.988 | 1.23 (0.66-1.90) | 0.667 | 0.84 (0.43-1.62) | 0.607 |
| Hypertriglyceridemia | 1.15 (0.74-1.78) | 0.515 | 1.32 (0.76-2.29) | 0.322 | 1.25 (0.72-2.15) | 0.417 |
| Hypercholesterolemia | 1.26 (0.65-2.44) | 0.486 | 1.66 (0.59-4.64) | 0.330 | 0.85 (0.34-2.10) | 0.725 |
| LDL-C ≥130 mg/dL | 1.30 (0.36-4.67) | 0.682 | 3.58 (0.39-35.51) | 0.227 | 0.56 (0.09-3.44) | 0.533 |
| Low HDL-C | 0.68 (0.18-2.56) | 0.579 | 1.011 (0.045-2.22) | 0.979 | 0.42 (0.03-4.74) | 0.475 |
| Conditiona | Dominant model for the XDH (518T>C) polymorphism TC+CC vs. TT | |||||
| All participants | Men | Women | ||||
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |
| Overweight and obesity | 0.97 (0.68-1.38) | 0.886 | 1.17 (0.68-1.99) | 0.559 | 0.85 (0.53-1.36) | 0.503 |
| SP ˃130 mmHg | 0.74 (0.24-2.23) | 0.594 | 0.85 (0.22-3.27) | 0.882 | 0.60 (0.08-4.23) | 0.609 |
| DP ˃85 mmHg | 0.95 (0.15-5.78) | 0.964 | 0.68 (0.04-11.13) | 0.791 | 1.2 (0.10-13.50) | 0.876 |
| Fasting glucose ˃100 mg/dL | 4.5 (0.55-37.23) | 0.122 | 1.38 (0.12-15.49) | 0.790 | 3.0 (0.35-26.63) | 0.284 |
| Hyperuricemia | 0.90 (0.63-1.34) | 0.613 | 0.77 (0.45-1.31) | 0.342 | 1.29 (0.64-2.61) | 0.470 |
| Hypertriglyceridemia | 0.84 (0.54-1.30) | 0.444 | 0.68 (0.39-1.18) | 0.174 | 1.41 (0.80-2.51) | 0.230 |
| Hypercholesterolemia | 1.30 (0.65-2.59) | 0.451 | 1.28 (0.46-3.60) | 0.630 | 1.44 (0.53-3.85) | 0.464 |
| LDL-C ≥130 mg/dL | 1.50 (0.38-5.87) | 0.555 | 1.03 (0.17-6.31) | 0.969 | 2.4 (0.27-22.15) | 0.411 |
| Low HDL-C | 1.28 (0.31-5.18) | 0.725 | 0.722 (0.350-1.70) | 0.522 | 1.8 (0.18-17.75) | 0.598 |
Abbreviations: BMI, body mass index; SP, systolic pressure; DP, diastolic pressure. LDL-C, LDL cholesterol; HDL-C, HDL cholesterol; ABCG2, gene that encodes an ATP-binding cassette transporter subfamily G member 2; SLC22A12, gene that encodes an urate transporter 1 gene; XDH, gene that encodes a xanthine dehydrogenase.
aThe cut-off criteria were according to The World Health Organization and the National Cholesterol Education Program Adult (ATP III). Overweight was defined as BMI ≥25, while Obesity was BMI ˃30; Hyperuricemia was defined as serum uric acid ˃6 and 7 mg/dL for women and men, respectively; hypertriglyceridemia as fasting serum triglycerides ˃150 mg/dL, hypercholesterolemia as fasting serum cholesterol ˃200 mg/dL, and low HDL-C as a serum concentration of ˂50 mg/dL and 40 mg/dL for women and men, respectively.
Multivariate logistic regression analysis: data are shown as odd ratio (95% Confidence Interval). The level of significance was set at p <0.05.
A total of 860 college applicants between the ages of 18 and 25 years of both sexes were included in this study, an important age range to identify early disorders. Indeed, we found a 15% prevalence of hyperuricemia, being higher among males (19.6%) than females (10%), as previously observed in another study with comparable age groups in Mexico (Alegría-Díaz et al., 2018). Regarding uric acid levels, similar blood concentrations have also been reported in Mexican young persons (Pérez-Navarro et al., 2020). The subgroup with hyperuricemia was prone to higher levels of BMI, blood pressure, triglycerides, total cholesterol, and LDL cholesterol (Table 1). In fact, the association between serum uric acid levels and the traits of metabolic syndrome has been previously analyzed (Lin et al., 2006; Zhang et al., 2020a), although some authors found no causal evidence of uric acid levels being associated with metabolic syndrome and its components (Wang et al., 2020). According to our results, being a man, being overweight or obese, or having dyslipidemia are related to high uric acid levels (Figure 1). Men have a lower capacity to eliminate urate via the kidney compared with females due to a deficiency of estrogen and progesterone (Hak et al., 2010). Likewise, the involvement of uric acid in lipid metabolism can lead to hyperuricemia, a condition considered as predictor factor of dyslipidemia (Kuwabara et al. 2020; Lima et al., 2015; Son et al., 2016) and can subsequently alter blood pressure (Teng et al., 2011; Zhang et al., 2020a). In this way, our results summarized in Figure 1 show that being male, being obese and having dyslipidemia increases the risk of hyperuricemia (Liu et al., 2020a).
The study of the genetic influence on blood uric acid levels has included the search for single nucleotide polymorphisms (SNP) in genes involved in purine metabolism and urate removal. Here, we analyzed three polymorphisms in three different genes, including ABCG2 (Q191K), SLC22A12 (517G>A), and XDH (518T>C). The first corresponds to the exchange of a glutamine for a lysine at position 141 of an adenosine triphosphate (ATP)-binding cassette transporter subfamily G member 2 (ABCG2). This exchange predisposes individuals to hyperuricemia (Nakashima et al., 2020; Wrigley et al., 2020). A frequency of 0.25 for the risk for T allele was found, a slightly lower prevalence than that reported in Asian and New Zealand populations (Kim et al., 2015; Liu et al., 2020b; Toyoda et al., 2019). The distribution of the T allele was marginal between hyperuricemic and normouricemic groups, being more frequent in the hyperuricemic group (p = 0.056). When a dominant model was carried out, the risk for the T allele was associated with hyperuricemia and hypercholesterolemia (total and HDL cholesterol). This association was confirmed only in men, showing that the ABCG2 (Q191K) polymorphism increases the risk for hyperuricemia 2.43 times (95% CI: 1.41-4.17, p = 0.001) and hypercholesterolemia 4.89 times (95% CI: 1.54-15.48, p = 0.003) in a dominant model. Other studies have also found a greater influence of this polymorphism in men (Narang et al., 2019) although, under some conditions, the ABCG2 (Q191K) polymorphism could contribute to increased uric acid in women (Guo et al., 2020; Roman et al., 2020). Although initially our work focused on hyperuricemia in young people, the increase in serum cholesterol in men associated with the ABCG2 (Q191K) polymorphism was observed. There are still few studies linking the ABCG2 gene and cholesterolemia. A relevant fact is that mRNA expression levels of ABCG2 appear to be higher in individuals with hypercholesterolemia (Rodrigues et al., 2009); likewise, the activity of ABCG2 has been associated with cholesterol levels both in vitro and in vivo (To et al., 2014). Therefore, perform genotype-based mRNA expression analysis to further explore the role of the ABCG2 (Q191K) polymorphism in hyperuricemia and hypercholesterolemia should be contemplated.
The SLC22A12 (517G>A) polymorphism comprises the transition from guanine to adenine in the 3´-UTR region in the urate transporter 1 gene; this transition appears to modify uric acid levels (Flynn et al., 2013; Köttgen et al., 2013). In this study, the frequency of the minor allele was 0.32; however, there was no statistically different distribution of this between hyperuricemic and normouricemic groups. Although no differences in uric acid levels were associated with this polymorphism, lower LDL cholesterol levels were observed in the group of carriers of the A allele when data were analyzed in a dominant model (p = 0.043), suggesting a protective effect of this allele (Simon et al., 2014). Since the SLC22A12 (517G>A) polymorphism is located at a potential miRNA binding site (Flynn et al., 2013), the epigenetic mechanism involved in the regulation of uric acid levels needs to be studied.
With respect to the XDH (518T>C) polymorphism, we studied the xanthine dehydrogenase polymorphism located in the 3´-UnTRanslated (UTR) region of the XDH gene. The replacement of T by C is considered a risk factor related to hypertension (Wu et al., 2015). We found that the minor allele frequency was 0.37 for the C allele, contrary to what was reported in a Chinese population, where the C allele had a higher frequency, in addition to its being associated with hypertension (Wu et al., 2015). However, the link between the XDH (518T>C) polymorphism and hypertension was not demostrated in Taiwanese women (Lee et al., 2019). In the present study, we did not find a statistically significant different distribution of the C allele between the hyperuricemic and normouricemic groups, and the risk allele was also not associated with any component of the metabolic syndrome.
Finally, hyperuricemia is an undesirable condition that has been seen as a minor trait of metabolic syndrome, cardiovascular risk, as well as other types of disorders such as psoriasis and alopecia, which have been associated with high levels of blood uric acid (Guo et al., 2020; Ma et al., 2020; Talebi et al., 2020; Zhang et al., 2020a). Even though our criterion for hyperuricemia were defined as blood uric acid >6 mg/dL for women and 7 mg/dL for men, cut-off points could be reconsidered according to the considerations made by Alegría-Díaz et al. (2018). In this regard, a recent study considers uric acid levels below 5 mg/dL for men and below 2-4 mg/dL for women to be optimal for a lesser risk of cardiometabolic diseases in a Japanese population (Kuwabara et al., 2020). Although there are modifiable factors related to lifestyle, genetic inheritance plays a decisive role in the control of uricemia.
This study has three main limitations: i) only one polymorphism was genotyped for every gene, analysis of multiple SNPs by haplotypes could be more appropriate; ii) the analysis of the ABCG2 (Q191K) polymorphism is limited by the small sample size and the compromised statistical power; and, iii) new cut-off values for hyperuricemia have been suggested (Alegría-Díaz et al., 2018); however the conservative criteria of >6 mg/dL for women and 7 mg/dL for men were considered in this study.
In this study, we found that the ABCG2 (Q191K) polymorphism increases the risk of hyperuricemia as well as of hypercholesterolemia in young Mexican males. Since the ABCG2 (Q191K) polymorphism can modify the efficacy of statins in reducing cholesterol (Zhang et al., 2020b), carriers of the risk allele represent a vulnerable group of interest for future pharmacogenetic research. Some considerations for future studies are including lifestyle and diet factors, the monitoring of the study population, and exploring more polymorphisms in the ABCG2, SLC22A12, and XDH genes, as well as studying haplotypes.
Data mendeley: Polymorphisms of the genes ABCG2, SLC22A12 and XDH and their relation with hyperuricemia and hypercholesterolemia in Mexican young adults. http://dx.doi.org/10.17632/243ft29b7m.1 (Alegría-Torres, 2021).
This project contains the following underlying data:
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular Epidemiology.
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. He J, Wang MY, Qiu LX, Zhu ML, et al.: Genetic variations of mTORC1 genes and risk of gastric cancer in an Eastern Chinese population.Mol Carcinog. 2013; 52 Suppl 1: E70-9 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular Epidemiology.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Metabolic syndrome, hyperuricemia, insulin resistance, endocrinology, internal medicine, urology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 24 Sep 21 |
read | |
|
Version 1 17 Mar 21 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)