Keywords
Calcium Imaging, Denoising, Motion Correction, Classification, Quantification, Machine Learning, Neural Networks
This article is included in the NEUBIAS - the Bioimage Analysts Network gateway.
This article is included in the Bioinformatics gateway.
This article is included in the Artificial Intelligence and Machine Learning gateway.
Calcium Imaging, Denoising, Motion Correction, Classification, Quantification, Machine Learning, Neural Networks
This latest version of the review article "Calcium imaging analysis – how far have we come?" contains a clearer breakdown of the possible image analysis options for different experimental set-ups of calcium imaging experiments such as in vitro versus in vivo methods. We also expand on the 'Quantification' section as this is an important section that was previously very brief. In this section we discuss identification of action potentials from calcium signals as this can be a controversial topic.
See the authors' detailed response to the review by Christopher Rowlands and Isabell Whiteley
See the authors' detailed response to the review by William Zeiger
See the authors' detailed response to the review by Anubhuti Goel
The ability to image calcium ion (Ca2+) dynamics in cells has long been of interest, particularly in the neurosciences, where it can be used as a marker for neuronal excitability. The origins of calcium imaging began in the mid-1970s (Blinks et al., 1976; Moisescu et al., 1975), however the most Ca2+ specific BAPTA (1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid)-based dye was developed in 1980 by Roger Tsien, and its derivatives are still used today (Tsien, 1980). In the past forty years, the methods available for measuring Ca2+ fluxes in cells have expanded to include ratiometric, fluorescence lifetime, or fluorescence intensity-based dyes, and genetically-encoded calcium indicators (GECIs) (Miyawaki et al., 1997; Ohkura et al., 2005). The use of microscopy modalities has also advanced to include light-sheet microscopy (LSM; Huisken et al., 2004) for long-term imaging, and 2-photon microscopy (2PM; Denk et al., 1990) for deep tissue and cell specific uncaging techniques. The combination of Ca2+ indicator and imaging modality used will reflect the properties of the sample and the scientific question, as well as the methodologies available to the researcher. For example, in vitro imaging, or in vivo invertebrate imaging, may use exogenous or GECIs, imaged using LSM, epifluorescence, 2PM or other fluorescence microscopes depending on the temporal and spatial resolution, timescale of imaging, and thickness of the sample being taken into consideration. Other specialist options for in vivo imaging of GECIs are available for imaging in awake and behaving animals including miniaturized forms of 1- or 2- photon endoscopic fluorescence microscopes (miniscopes) for single-cell in vivo recordings (Cai et al., 2016; Chen et al., 2013; Silva, 2017).
Calcium imaging is an inherently noisy method due to the high spatiotemporal information desired from a sample often showing low signal-to-noise alongside drift or cell movement, particularly for living organisms. In recent years, a number of software packages have been written for individual aspects of the commonly used pipeline in calcium imaging analysis (Figure 1). This processing pipeline includes image denoising, motion correction, classification for cell identification, and quantification of calcium signals. As calcium imaging is used across a broad range of samples, from sub-cellular, cellular, networks, bulk tissue dynamics to whole organisms and behaving animals, aspects of this pipeline can vary substantially with no ‘one size fits all’ approach.
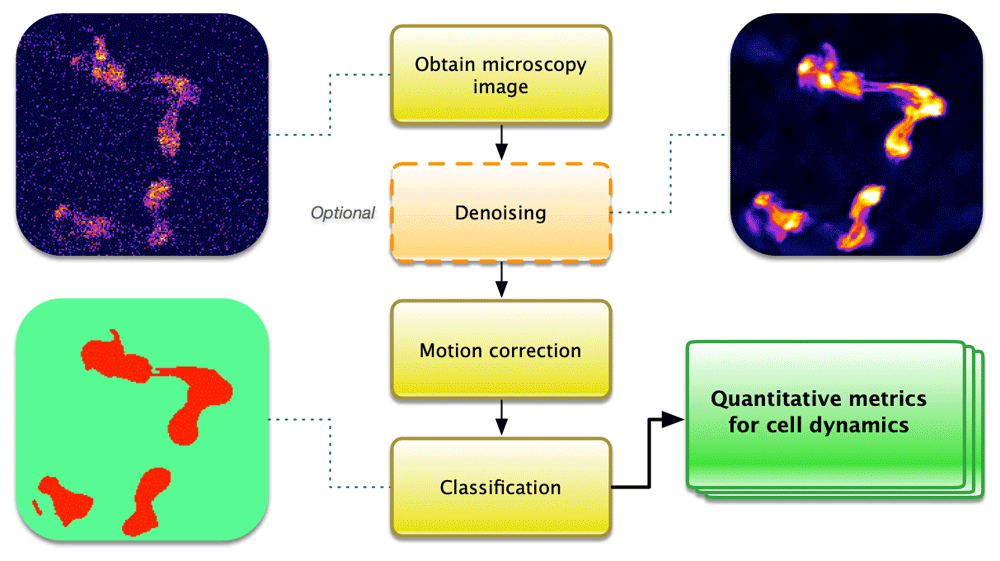
Denoising is an optional step that can help to improve signal-to-noise and enhance features. Motion correction may be necessary in cases of drift or movement. Classification can select regions of interest for which quantitative analysis is performed.
Live-cell imaging generally requires short exposure times and low excitation power to limit the effects of photo-toxicity and photo-bleaching. This leads to image degradation in the form of noise. In fluorescence microscopy the two prevalent noise sources are Poisson noise and Gaussian noise. Poisson noise is caused by the stochastic and discrete nature of photon emission and tends to be dominant at low light levels, whereas Gaussian noise describes the intrinsic thermal and electronic fluctuation in the image sensor (Luisier et al., 2011)
Although denoising is not a required step in the pipeline, effective denoising can improve the subsequent steps by artificially enhancing signal-to-noise. Traditionally, image denoising has been based on local averaging approaches, such as the application of a Gaussian smoothing filter (Buades et al., 2005; Lindenbaum et al., 1994). Alternative methods include a local filter method such as anisotropic filters (Broser et al., 2004; Kitamura & Häusser, 2011) (Perona & Malik, 1990), or in the frequency domain, Wiener filters (Wiener, 1950) and wavelet thresholding methods (Donoho, 1995)(Besbeas et al., 2004; Wegner et al., 2006).
Local methods are computationally light but have clear limitations. First, the averaging often involved in local methods introduces blur, causing features to appear less well defined. Second, they do not perform well for high noise levels, since the correlations between neighbouring pixels deteriorate (Shao et al., 2014). In the context of calcium imaging, local methods have been shown to perform well (Malik et al., 2011).
Non-local filters solve some of the problems by using self-similarity of natural images beyond neighbouring pixels, thus exploiting global information (Shao et al., 2014). The first method to propose this is the non-local means method (Buades et al., 2005), in which subregions of an image referred to as patches are restored by weighted averaging of all other patches in an image. Since then, there have been a number of improvements such as invariance to patches that are rotated or mirrored with respect to each other (Grewenig et al., 2011), improved computational efficiency, and automated parameter tuning and extension to 3D image stacks (Coupé et al., 2008). Although non-local filters are better at high noise levels, they will typically lead to artefacts like over-smoothing (Shao et al., 2014). A modern, well-balanced and state-of-the-art non-local method is ND-SAFIR, which is specifically geared towards application in fluorescence microscopy imaging (Boulanger et al., 2010). ND-SAFIR is a powerful method for removing Poisson-Gaussian noise. It is based on non-local means denoising using a variance stabilisation step, followed by calculating the spatial and temporal patch-based weighted averages for intensity values. The method is widely applicable between experimental samples and can be used directly for 2D+t and 3D+t datasets (Buades et al., 2011).
In recent years, deep learning methods have become state-of-the-art for denoising. Methods such as DnCNN (Zhang et al., 2017), FFDNet (Zhang et al., 2018) and CARE (Weigert et al., 2018) rely on convolutional neural networks that are trained in a supervised learning approach. However, this requires ground truths to be available for model training, which may be difficult to obtain in practice. A different approach was developed in noise2noise (Lehtinen et al., 2018), where instead of learning the mapping from noisy images to clean targets, the model is trained with other noisy images as targets. The images must be corresponding pairs displaying the same objects but with independent noise. Assuming the noise sources underlying the images have zero-mean distributions, the weights of the network will then converge during training to the same values as a network trained with clean targets because the noise that manifests in the weights cancels out. A more recent method, noise2void (Krull et al., 2019), aims to resolve this issue of needing ground truths, by using self-supervised learning. Here, the network is optimised to predict the value of each pixel from the values of neighbouring pixels in an image, thus requiring no separate ground truths. In another recent method, DeepInterpolation (Lecoq et al., 2020), the need for ground truth training data is avoided by treating the denoising task as a nonlinear interpolation problem. This assumes that the data have a sequential component, such that spatiotemporally overlapping features can be exploited. DeepCAD is a new deep self-supervised denoising method that reduces detection noise and thereby improve the signal-to-noise more than tenfold, which it claims can improve the accuracy of neuron extraction and spike inference (Li et al., 2021).
Motion correction is often required to ensure consistent image processing across a time stack. We distinguish between two types of motion: (a) drift occurring in the imaging system itself caused by thermal gradients in the microscope, vibrations and mechanical instability (Kreft et al., 2005); (b) subject motion such as fluctuations in the immersion media or the movement of organisms (Jenkinson et al., 2002). Drift will typically play a significant role when imaging the same field of view over multiple days, which can be rectified by using standard registration methods (Dubbs et al., 2016)(Thévenaz et al., 1998).
More complex motion such as organism movement can be harder to correct as it is often non-uniform, over a large area, and causes movement in-and-out of the focal plane. These require non-rigid registration methods or motion tracking. A commonly used example available in Python and MATLAB is Non-Rigid Motion Correction, NoRMcorre (Pnevmatikakis & Giovannucci, 2017), which uses patch-based field of view registration whereby separate images are then merged by smooth interpolation. The popularity of NoRMcorre may in part be due to its general applicability.
Two correction methods have been produced for in vivo imaging in awake rodents, one based on the rigid-transform-based Lucas–Kanade (gradient descent) (Lucas & Kanade, 1981) image registration algorithm using MathWorks® MATLAB platform (Greenberg & Kerr, 2009), the other using a Hidden Markov Model (Dombeck et al., 2007). Although effective, these methods have not been packaged for easy implementation and are reliant on cells remaining in the x- and y- dimensions as it cannot track following movement between z-axes. In cases with z-axis movement, tracking-based methods may be more reliable, and specialist options exist using control theory and machine learning approaches for post-processing (Nguyen et al., 2017), or applied to a motorised stage (Cong et al., 2017; Kim et al., 2017). A MathWorks® MATLAB toolbox, miniscope 1-photon imaging pipeline (MIN1PIPE), has been developed to include denoising, motion correction and signal extraction (Lu et al., 2018). MIN1PIPE motion correction includes several steps including the Lucas-Kanade and Kanade-Lucas-Tomasi (Lucas & Kanade, 1981; Shi & Tomasi, 1994) trackers, and Log-Demons registration (Vercauteren et al., 2009), and outperforms the Lucas-Kanade, Kanade-Lucas-Tomasi, and NoRMcorre for using sample 2-photon videos (Lu et al., 2018).
Tracking methods specifically designed to be more basic to implement and widely available include plug-ins for image processing packages (Abramoff et al., 2004) such as Trackmate (Tinevez et al., 2017), or Time Series Analyzer (Balaji, 2014).
Classification is required to ensure that the quantification can be performed over specific regions of interest, such as for subcellular area, specific cells, or tissue regions. Classification can be achieved through pixel- or object-based segmentation. Pixel-based methods map each pixel to a class according to the spectral similarities. Popular pixel-based methods for calcium image analysis include Maximum Likelihood Classification (MLC) (Malik et al., 2011) or Otsu thresholding to separate ‘light’ and ‘dark’ clustered pixels (Otsu, 1979) as used as part of the SIMA Python package ROI pipeline (Kaifosh et al., 2014).
Object-based segmentation is a two-step process using both spectral and spatial/contextual information to group pixels into objects which are then classified. CaImAn is an open-source package with modules for classification, motion correction, source extraction, and spike deconvolution. The classification method is based on convolutional neural networks (Giovannucci et al., 2019). It was packaged into EZcalcium in an effort to improve usability by providing a GUI in MathWorks® MATLAB (Cantu et al., 2020). However, using limited CaImAn function in EZcalcium does not easily allow for segmentation of more complex structures or large organelles or clusters of cells and is better for somas or smaller, less complex areas. Cellpose is another generalist, deep learning-based segmentation method that uses entirely open source packages in Python with a GUI to aid implementation. There is also a web-based option for testing Cellpose, which makes it very easy to use (Stringer et al., 2020), though it too can be limited at detecting more complex cell shapes such as dendrites and axons.
DenoiSeg is an extension of Noise2Void that offers an end-to-end neural network, which is jointly optimised to denoise and segment images. The denoising capability is learnt by the self-supervised learning principle that noise2void introduced (Krull et al., 2019). By combining this with a supervised learning approach using a few annotated ground truths of segmentation maps, the final segmentation performance ends up performing better than without co-learning, i.e. having two separate networks perform the respective tasks (Buchholz et al., 2020).
Cell classification methods have been discussed with the conclusion that ‘learning-based methods score among the best-performing methods, but well-optimized traditional methods can even surpass these approaches in a fraction of the time’ (Vicar et al., 2019).
The aim of each step is for signal extraction to allow a quantitative output from the images of calcium signals. The most commonly used measure is the relative fluorescence variation (ΔF/F0) for classified cells. Packages will therefore either provide this data of the baseline fluorescence (F0) and deviations from baseline (ΔF), for further analysis, or provide a direct plot. Background subtraction may need to be considered as not all packages will take this into account. Multiple methods can be used, including subtracting the intensity values from a region of the image that does not contain Ca2+ indicator from the intensity values in regions of interest. However care should be taken using background subtraction with ratiometric indicators (Shkryl, 2020). F0 baseline values can be calculated by averaging the values before the onset of stimulation in the same region (Galizia et al., 1999), or by low-pass filtering the signal (Balkenius et al., 2009) (For review (Balkenius et al., 2015).
Signal extraction from single cells can be particularly difficult for in vivo recordings due to large background fluxes and high spatial overlaps of cells outside of the focus plane which is further increased in 1-photon compared to 2-photon imaging. Semi-automated ROI analysis (Barbera et al., 2016; Klaus et al., 2017; Pinto & Dan, 2015), principal component analysis independent components analysis (PCA/ICA) (Mukamel et al., 2009), clustering based approaches (like Suite2P; (Pachitariu et al., 2017), and constrained nonnegative matrix factorization (CNMF) (Pnevmatikakis et al., 2016) approaches are techniques that have been explored with different strengths for detecting background and spatial overlap. An ‘extended’ CNMF method (CNMF-E) has been developed with an adjusted spatiotemporal background model that outperformed PCA/ICA for the simulated and experimental datasets that were tested (Zhou et al., 2018). For a package method, the toolbox MIN1PIPE combines a CNMF (Pnevmatikakis et al., 2016) with additional steps to remove false positives (Lu et al., 2018). CaImAn also builds upon the CNMF algorithm (Pnevmatikakis et al., 2016) to allow it to be fully automated, and CNMF-E for 1-photon endoscopic data (Zhou et al., 2018).
Another feature commonly needed by researchers is timing of neuronal action potentials (APs) or ‘spike detection’ through deconvolution of the extracted signal. A wide range of algorithms can be used as discussed in the results to the Spikefinder challenge (Berens et al., 2018) as there are multiple methods of varying complexity that can be used. EZcalcium directly shows the raw fluorescence, inferred activity and deconvolved neural ‘spiking’, whereby the data can then be exported into file formats for proprietary (.mat, .xlsx) or open (.csv) software programmes for further analysis (Cantu et al., 2020; Giovannucci et al., 2019). The ability to accurately detect spikes requires knowledge of ground truth, usually from electrophysiological recordings. Calcium imaging can be susceptible to variation between neuron type, calcium indicator and its concentration used, the optical resolution, the sampling rate and the noise level. Therefore, it is fundamental to understand how specific indicators react under the given imaging conditions, which cannot be readily generalized across protocols. To try and improve the accuracy of spike detection, a toolkit using a supervised algorithm of spike inference has been developed using a ‘ground truth database’ from a large number of sets of calcium imaging with corresponding electrophysiological measurements (Rupprecht et al., 2021).
A great number of analysis advancements have been made since calcium imaging was first developed. Popular packages for various steps of the pipeline (Figure 1) include CaImAn, SIMA, Suite2P, and EZcalcium (Cantu et al., 2020; Giovannucci et al., 2019; Kaifosh et al., 2014; Pachitariu et al., 2017). Although these packages are great starting tools for the community, many require programming knowledge in Python or commercial packages such as MathWorks® MATLAB. Many of the available options are only semi-automated and the limited automated options available are often designed for a very limited experimental context and are not actively supported when problems are experienced, e.g. other than for cells of a specific size and shape imaged in vitro. EZcalcium is one of the most intuitive options, which has improved the usability of CaImAn, NoRMCorre, but again seems best suited to analyse cell bodies. Suite2P and EZcalcium both attempt to offer an automated pipeline from raw images to spike extraction with no prior programming knowledge required by the user (Cantu et al., 2020; Pachitariu et al., 2017). As both packages are suited to similar experimental data, the choice may be based upon personal preference.
It therefore seems that perhaps some of the biggest advances could be made by designing packages for detecting neuritic structures or organelles and improving the spatial resolution of the analysis to be intracellular, such as has been used for calcium sparks (Berens et al., 2018). Longitudinal tracking of specific cells across imaging sessions also remains a challenge so that individual cells can be identified between multiple imaging sessions. A MathWorks® MATLAB toolkit has been made with reported error rates of < 5 % (Sheintuch et al., 2017); an alternative approach is also available using CaImAn (Giovannucci et al., 2019) though direct comparisons between these methods is difficult without knowing ground truths. Calcium imaging for population activity has also been highlighted as an area that requires further research, particularly when imaging over larger fields of view. Using models specific for neuron types imaged may improve detection of APs, which are commonly under-represented in population activity measurements (Huang et al., 2021). Recent toolboxes with large datasets containing ground-truths may reduce false negatives during analysis (Rupprecht et al., 2021). On the other end of the scale, pipelines for functional imaging in organisms such as zebrafish, C. elegans and Drosophila, where motion correction is often required and improved analysis for connectomics purposes are much needed.
As the application of machine learning in calcium imaging analysis matures, a higher level of automation and throughput for analysis tasks can be expected to follow. This will be enabled by more generalised and robust machine learning models. The barrier to training and deploying these methods will also reduce as more research is made into few-shot learning (using small training datasets) in addition to training approaches such as self-supervised and unsupervised learning.
No data are associated with this article.
This publication was supported by COST Action NEUBIAS (CA15124), funded by COST (European Cooperation in Science and Technology).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Systems neuroscience, Calcium imaging, mouse behavior, electrophysiology.
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Microscopy, instrument development, automation and data analysis, neurophotonics, biophotonics
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Stroke, neural circuits, two-photon calcium imaging
Is the topic of the review discussed comprehensively in the context of the current literature?
Yes
Are all factual statements correct and adequately supported by citations?
Partly
Is the review written in accessible language?
Yes
Are the conclusions drawn appropriate in the context of the current research literature?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Systems neuroscience, Calcium imaging, mouse behavior, electrophysiology.
Is the topic of the review discussed comprehensively in the context of the current literature?
Partly
Are all factual statements correct and adequately supported by citations?
Yes
Is the review written in accessible language?
Yes
Are the conclusions drawn appropriate in the context of the current research literature?
Yes
References
1. Rupprecht P, Carta S, Hoffmann A, Echizen M, et al.: Database and deep learning toolbox for noise-optimized, generalized spike inference from calcium imaging. bioRxiv. 2020. Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Stroke, neural circuits, two-photon calcium imaging
Is the topic of the review discussed comprehensively in the context of the current literature?
Yes
Are all factual statements correct and adequately supported by citations?
Yes
Is the review written in accessible language?
Partly
Are the conclusions drawn appropriate in the context of the current research literature?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Microscopy, instrument development, automation and data analysis, neurophotonics, biophotonics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 26 Aug 21 |
read | read | read |
|
Version 1 30 Mar 21 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)