Keywords
COVID-19, SARS-CoV-2, HCV, HIV, Ebola, Zika, PubMed, Scientometrics
This article is included in the Emerging Diseases and Outbreaks gateway.
This article is included in the Research on Research, Policy & Culture gateway.
This article is included in the Coronavirus (COVID-19) collection.
COVID-19, SARS-CoV-2, HCV, HIV, Ebola, Zika, PubMed, Scientometrics
We expanded the introduction section with Table 1, which compares five viruses examined in the paper.
We examined the types of the publications in COVID-19 research according to the publication-classification scheme of PubMed.
We discussed the limitations of our approach.
See the authors' detailed response to the review by Mahmoud Nassar
See the authors' detailed response to the review by Ludovico Abenavoli
The recent outbreak of coronavirus disease 2019 (COVID-19) has imposed an unprecedented and devastating burden on the world,1 including a serious encumbrance to health care systems.2 Collectively the scientific community has responded to the pandemic by researching the spread of the disease and its causative pathogen, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), in order to understand and terminate the pandemic. These efforts have resulted in a vast amount of publications. We believe it would be worthwhile to analyze the trend of the publications in order to predict the future of research in this area.
We have previously demonstrated that the number of publications may be a reliable quantitative measure of the magnitude of research activity of a biological or biomedical science.3 In conjunction with regression analysis, the method of assessing research activity of a biological or biomedical discipline based on the number of publications in the field has been found to be effective in the prognostication of the future of biomedical fields by extrapolation of the best fit equation.4 The method has successfully been applied to various fields such as food sciences,5 epigenetics,6 metabolomics,7 and environmental sciences.8
In this paper, we apply the method mentioned above to COVID-19 research to quantitatively describe the temporal development of the research and predict its future. We also include four other viruses in the study; hepatitis C virus (HCV) and HIV as negative controls without any apparent outbreaks in the period from January to November, 2020, and Ebola virus disease (EVD) and Zika virus (ZIKV) as positive controls of epidemiological outbreak during the period of examination from January 2014 to November 2020. Comparison of those five viruses is in Table 1. One thing noteworthy to mention is the relatively big genome size and small GC content of SARS-CoV-2 (Table 1).
| Virus | RefSeq | INSDC | Size (Kb) | GC% | Protein | Gene | URL* |
|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | NC_045512.2 | MN908947.3 | 29.9 | 38 | 12 | 11 | 1 |
| HCV genotype 1 | NC_004102.1 | AF009606.1 | 9.65 | 58.2 | 2 | 1 | 2 |
| HCV genotype 7 | NC_030791.1 | EF108306.2 | 9.44 | 56.8 | 2 | 1 | 3 |
| HIV 1 | NC_001802.1 | AF033819.3 | 9.18 | 42.1 | 10 | 10 | 4 |
| Ebola | Not available | LT605058.1 | 18.96 | 41.2 | 9 | 7 | 5 |
| Zaire ebolavirus | NC_002549.1 | AF086833.2 | 18.96 | 41.1 | 9 | 7 | 6 |
| Zika | NC_012532.1 | AY632535.2 | 10.79 | 50.9 | 1 | 1 | 7 |
* URL
1. https://www.ncbi.nlm.nih.gov/genome/?term=sars-cov-2%5Borgn%5D
2. https://www.ncbi.nlm.nih.gov/genome/?term=HCV%5Borgn%5D
3. https://www.ncbi.nlm.nih.gov/genome/?term=HCV%5Borgn%5D
4. https://www.ncbi.nlm.nih.gov/genome/10319
5. https://www.ncbi.nlm.nih.gov/genome/4887?genome_assembly_id=414173
6. https://www.ncbi.nlm.nih.gov/genome/4887?genome_assembly_id=888630
7. https://www.ncbi.nlm.nih.gov/genome/?term=zika+virus%5Borgn%5D
To quantitatively investigate the trend of research related to the five viruses (SARS-CoV-2, HCV, HIV, EVD, and ZIKV), we searched the PubMed database on December 23, 2020. Our search strategy was as follows for the different viruses: (The superscripts a and b in the search phrases represent month and year, respectively.)
SARS-CoV-2: (((COVID [Title/Abstract]) OR (COVID-19[Title/Abstract])) OR (SARS-CoV-2[Title/Abstract])) AND ((“2020/Ma”[Date - Publication]: “2020/Ma”[Date - Publication]))
HCV: (((HCV [Title/Abstract]) OR (“hepatitis C virus”[Title/Abstract])) AND (virus [Text Word])) AND ((“2020/Ma”[Date - Publication]: “2020/Ma”[Date - Publication]))
HIV: (((HIV [Title/Abstract]) OR (“human immunodeficiency virus”[Title/Abstract])) AND (virus [Text Word])) AND ((“2020/Ma”[Date - Publication]: “2020/Ma”[Date - Publication]))
Ebola: ((Ebola [Title/Abstract]) AND (virus [Text Word])) AND ((“Yb/Ma”[Date - Publication]: “Yb/Ma”[Date - Publication]))
Zika: ((ZIKA [Title/Abstract]) AND (virus [Text Word])) AND ((“Yb/Ma”[Date - Publication]: “Yb/Ma”[Date - Publication]))
The number of publications on each virus was manually recorded on a monthly basis for eleven months for SARS-CoV-2, HCV, and HIV from January to November 2020, and for eighty-three months for EVD and ZIKV from January 2014 to November 2020 for further investigation of data. Subsequent nonlinear regression analysis of the PubMed search results was conducted to obtain equation of best fit using SigmaPlot (version 11; Systat Software, Inc., San Jose, CA).
We retrieved monthly publication numbers of the five viruses from the Pubmed database, and obtained the best fitting equation for each virus. Our results are summarized in Figure 1 and Table 2. Underlying raw data of our research are available in the database of Figshare.9 We identified that temporal dynamics of publications related to the five viruses exhibit four characteristics.
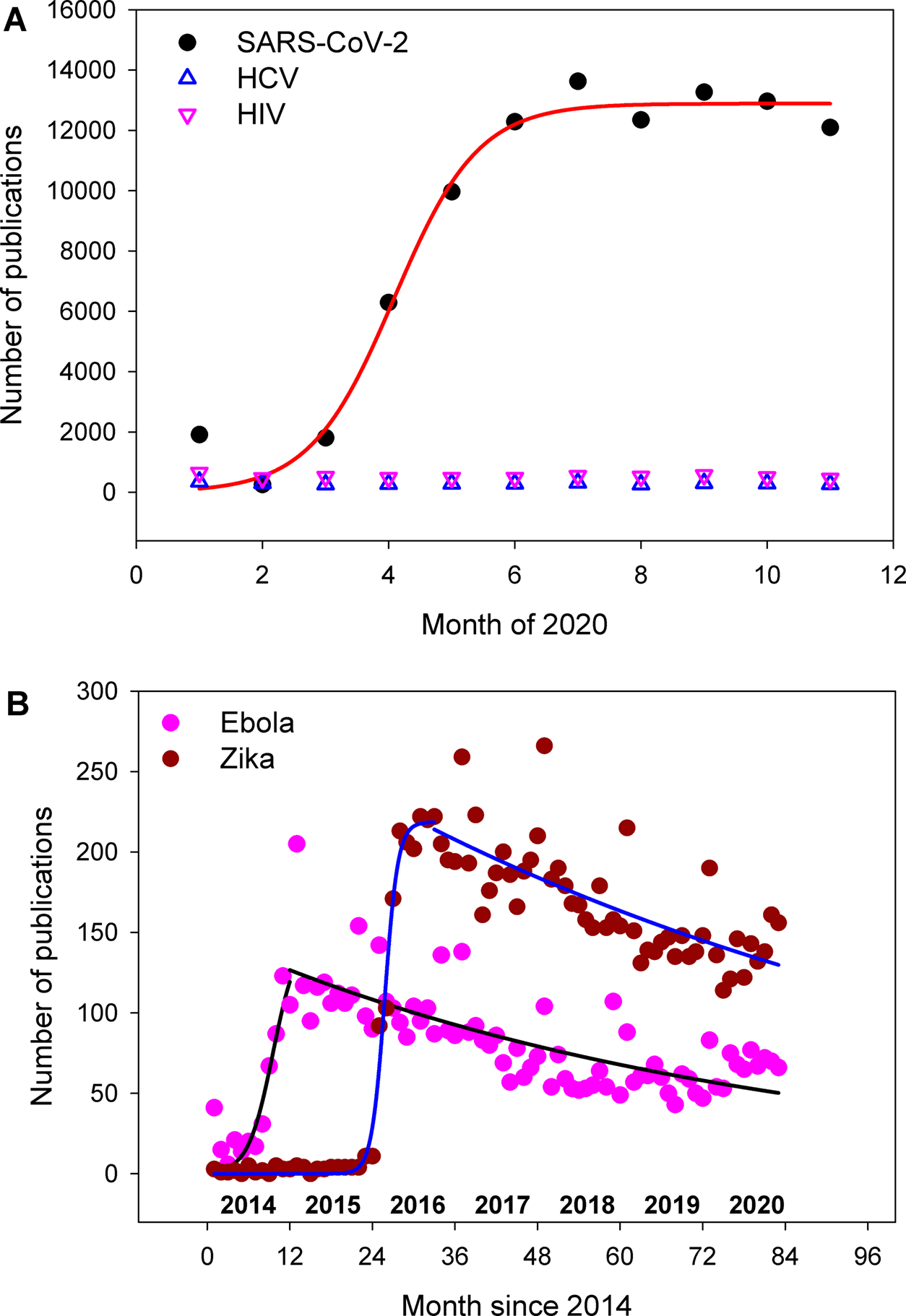
The solid line in each graph represents the best fit. The corresponding year in the panel B is presented above the x-axis. SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2.
| Equation | Virus | a ± SE | b ± SE | c ± SE | R2 |
|---|---|---|---|---|---|
| Eq. (1) | SARS-CoV-2 | 12900 ± 370 | 0.67 ± 0.12 | 4.1 ± 0.14 | 0.9803 |
| Ebola | 150 ± 65 | 1.8 ± 0.87 | 9.5 ± 1.9 | 0.8345 | |
| Zika | 220 ± 3.9 | 0.96 ± 0.088 | 25.8 ± 0.1 | 0.9916 | |
| Eq. (2) | Ebola | 150 ± 8.7 | 0.013 ± 0.001 | 0.5522 | |
| Zika | 300 ± 22 | 0.01 ± 0.001 | 0.5466 |
First, a sigmoidal equation (Equation 1) was found to be the best quantitative description of the publication trend of COVID-19 research:
The value of each parameter is listed in Table 2. The mathematical meaning of each parameter can be found in our previous publication.4 In brief, the parameter “a” represents an asymptotic maximum value of the function, “b” is related to the shape of the function, and “c” is the year when the value of the function is half of the asymptotic maximum value.4 The sigmoidal kinetics observed in the research trend of COVID-19 (Figure 1) is congruent with other areas of research such as bioinformatics, epigenetics, food sciences, and environmental sciences.4–7
Second, there was no significant correlation between the temporal point and the number of research publications on HCV and HIV during the time period examined from January to Novmber 2020 (p = 0.240 for HCV, and p = 0.367 for HIV) (Figure 1). This can be attributed to the absence of any significant outbreaks of HCV or HIV during the time period; while these viruses are important in a biomedical sense,10,11 those viruses have likely been endemic.12,13
Third, two examples of outbreaks in the decade of 2010, EVD14 and ZIKV,15 exhibit biphasic kinetics in the publication trend (Figure 1). The phase of sharp increase in number of publications, which overlaps with the time of each outbreak, also follows sigmoidal kinetics (Equation 1 and Table 2) as does COVID-19. The second phase, a decreasing phase, shows a slow and gradual decline that can be described by an exponential decay function (Equation 2):
Fourth, the exponential nature of the decay kinetics may be valuable for the prediction of the future of COVID-19 research. In the case of EVD, the publication number started to decrease, when x = 11 (Figure 1), where the publication number is 123 (see underlying data9) corresponding to 82% of the asymptotic maximum value of 150 (Table 2). Zika research started to decrease, when x = 33 (Figure 1), where the publication number is 222 (see underlying data9) corresponding to 101% of its asymptotic maximum value of 219 (Table 2). As of June, 2020, COVID-19 research reached 95% of its asymptotic maximum value of 12900 (Figure 1): 12288/12900 = 0.95 (underlying data9 and Table 2). The quantitative comparison between SARS-CoV-2 and the two viruses clearly suggests that the case of ZIKV is a more appropriate model for the prediction of COVID-19 research. Despite the apparent similarity of the research trend between SARS-CoV-2 and ZIKV, one should note that there is a substantial difference in the asymptotic maximum value (a in Equation 1) between these two areas of research: SARS-CoV-2 has an almost 60 times (≅ 12900/220) larger value of a than ZIKV (Table 2).
In addition to quantitative analysis of the publication trend of COVID-19 research, we examined the types of the publications according to the publication-classification scheme of PubMed. While most publications belong to the category of journal article, COVID-19 related research has been published in various other formats of publication. Complete data is available in the database of Figshare.16
In this study, we examined trends of publications related to five viruses focusing on SARS-CoV-2 using the PubMed database. Reviewer 2 to our initial version raised a question on our selection of PubMed in literature search rather than MEDLINE or Embase.17 Clarification to this comment may be useful. MEDLINE is the National Library of Medicine’s bibliographic database and is the primary component of PubMed.18 Embase is a commercial database of biomedical research (https://www.elsevier.com/solutions/embase-biomedical-research). While it can be useful in biomedical literature search, paying charges is required to access the database. In contrast, PubMed is a free resource for biomedical literature search (https://pubmed.ncbi.nlm.nih.gov/about/), and has been found effective in our previous research.3-8
The results of our research have implications for three sectors of the global community. One is for the scientific community in that research on COVID-19 is predicted to be active for a long time, even after commencing a downward trend. According to our mathematical model of the research on ZIKV, it will take COVID-19 research approximately 5 years (65.8 months) to reach half of its maximum value: f2(98.8) = f1(33)/2 and 98.8 – 33 = 65.8. While it is not certain when the publications on COVID-19 will start to decline, we expect that it will remain a major topic of research until at least 2025. This prediction may serve as a guide in planning research on COVID-19. The second implication of our results is for researchers in epidemiology as the method introduced in this paper can be easily applied to other epidemics and pandemics. The third implication is for young students. Our analysis of the ongoing research on COVID-19 should show them that science is a valuable way of contributing to humanity by providing solutions for public concerns such as COVID-19.
Finally, we conclude our study with limitations of our study. The fundamental rationale of our study is the future may be predicted by analyzing the history, which is one of the approaches used in the research of complex systems.19 While this approach was proved to be effective in predicting the future trend in our previous study,4 the future is intrinsically uncertain, especially when it involves human behaviors.20 For example, the number of publications in biochemistry showed a sudden increase between 1974 and 1975, which was far out of regularity.3 The most conceivable factor that may limit our prediction is emergence of novel disastrous mutants of SARS-CoV-2. Another factor would be the effectiveness or medical implications of vaccines and therapeutic agents for COVID-19.21,22 It will be interesting to evaluate the validity of our prediction made in this paper in 2025.
Figshare: Number of PubMed-indexed articles related to five viruses; SARS-CoV-2, HCV, HIV, Ebola, and Zika. https://doi.org/10.6084/m9.figshare.12958361.v39
This project contains the following underlying data:
- covid_figshare_kang.csv (spreadsheet of the number of research publications found relating to five viruses).
Figshare: Number of COVID-related publications in each publication type according to PubMed. https://doi.org/10.6084/m9.figshare.17283764.v516
This project contains the following underlying data:
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
We would like to thank Kathi Canese, a Program Manager for the support department of PubMed, for the valuable advice in the search of PubMed.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bioinformatics, cardiovascular
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Nassar M, Nso N, Alfishawy M, Novikov A, et al.: Current systematic reviews and meta-analyses of COVID-19.World J Virol. 2021; 10 (4): 182-208 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Clinical research, COVID-19, Cardiovascular Diabetes, and Organ transplantation.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Epidemiology of COVID-19, COVID-19 clinical aspects, COVID-19 research trends
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 21 Apr 22 |
read | ||
|
Version 1 12 Apr 21 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)