Keywords
COVID-19, SARS-CoV-2, ACE2, Molecular Docking, Drug Repurposing
This article is included in the Pathogens gateway.
This article is included in the Emerging Diseases and Outbreaks gateway.
This article is included in the Cheminformatics gateway.
This article is included in the Coronavirus (COVID-19) collection.
COVID-19, SARS-CoV-2, ACE2, Molecular Docking, Drug Repurposing
Edits to the manuscript focus on an improved and better organization on the abstract, methods and discussion sections, accordingly to reviewers suggestions. New references were cited on manuscript to support Introduction and Discussion sections in order to addresses the reviewers comments.
See the authors' detailed response to the review by Yogendra Nayak and Krishnaprasad Baby
See the authors' detailed response to the review by Fernando Yepes-Calderon
Emerging viruses can be defined as those whose incidence has increased in the last twenty years or whose presence has a high probability of increasing in the near future. Diseases caused by emerging viruses are one of the biggest public health threats globally1. Some of the viruses that fall within this catalog are the avian influenza virus subtype H5N1, severe acute respiratory syndrome (SARS), Ebola, Zika, and MERS-CoV, to name a few2. Coronaviruses (CoVs) are classified into four genera, α-CoV, β-CoV, γ-CoV, and δ-CoV2. α and β infect mammals, γ birds and δ birds and mammals, respectively3. These viruses are of public health importance because they cause enteric, renal, and neurological respiratory diseases that range from asymptomatic to fatal4,5.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) appeared in Wuhan, China, in December 2019, causing cases of SARS-like atypical pneumonia6,7, with a clinical picture of fever, general malaise, dry cough, shortness of breath and was called the coronavirus disease 2019 (COVID-19)8. It can be asymptomatic, develop mild-to-severe symptoms, or may cause death in patients with chronic diseases, such as hypertension, diabetes, and obesity9. On January 31st 2020, the World Health Organization (WHO) declared COVID-19 a public health emergency of international concern and, on March 12th, it was declared a global pandemic10. In Mexico, local transmission (phase 2 of transmission) was declared on March 24th 2020, which resulted in the suspension of non-essential activities in the country, generating economic losses in addition to public health problems and deaths associated with the disease. As of March 1st 2021, Mexico had reached 2.1 million cases of COVID-19 and 186 thousand deaths; around 116 million cases and 2.57 million deaths have been reported worldwide.
To date, there is no specific commercial treatment to control the infection11–13. Measures such as early detection, blocking the route of transmission through social isolation, isolation of suspected cases, disinfection of objects, as well as frequent hand washing with soap, in addition to the use of biosafety equipment such as surgical masks for health personnel, may reduce the transmission of COVID-19 among population.
Coronaviruses, such as SARS-CoV-2, are positive-stranded RNA viruses enveloped on a membrane. The coronaviral genome is composed of approximately 30,000 nucleotides containing the envelope (E), membrane (M), spike (S), nucleocapsid (N) and ORFs, that encode non-structural proteins, including enzymes that appear during their in-host reproductive cycle-genes14.
This virus measures 70 to 100 nm and belongs to the genus β-CoV15 and it has been proposed that any of the aforementioned proteins that make up CoVs may be targets for the development of vaccines or drugs4. Protein S plays an essential role on COVID-19 infection as it mediates the internalization on host cell and for the spread of the virus in the infected host16,17. This starts when the receptor binding domain of the S1 subunit (RBDS1) of S protein binds to the peptidase domain of angiotensin-converting enzyme 2 receptor (ACE2)18 and it is know that disrupting the binding of S protein to ACE2 prevents the attaching an the later internalization of the virus to the host cell19.
This protein interaction has recently been crystallized and deposited in the Protein Data Bank database20, allowing us to use it as a model of study to test different strategies to counter SARS-CoV-2 infection, like blocking S glycoprotein-ACE2 interaction through the discovery of sites of potential pharmaceutical interest.
In 2019, Research and Development (R&D) spending in the pharmaceutical industry totaled 186 billion U.S. dollars globally and its projected to reach 233 billion U.S. dollars to 2026. Unfortunately, drug development takes large time and financial resources that not all countries possess, especially developing countries, like Mexico.
In this sense, drug repurposing or repositioning allow us to integrate all evidence, pharmacodynamics/kinetics, bioavailability, among other important parameters, from an existing and approved drug in order to manage emerging diseases, like COVID-19. All this translates into a considerable decrease in research time and investment of resources in R&D21.
Different approaches have been taken in order to disrupt SARS-CoV-2 protein S-ACE2 interaction, as an example, many works has focus on finding potential biding sites on protein S structure, however, new variant strains has been detected worldwide, like B117 in UK, P1351 in South Africa, P1 and P2 in Brazil. All variant strains display the N501Y mutation, which is located on the RBD of the S protein, making the interaction more effective. In this sense, targeting RBD may be a transitory approach, therefore, an alternative strategy would be aiming at the ACE2 receptor. Some authors have pointed out some concerns about using drugs that targeting the renin–angiotensin signaling (RAS) pathway22–24, but Jia and collaborators25 highlight current efforts of exploiting ACE2 as therapeutic target, like the use of pseudo-ligands to dominate the binding site for SARS-CoV-2 as an example. Therefore, inhibition of the SARS-CoV-2 protein S-ACE2 interaction trough aiming ACE2 receptor it is a plausible strategy. In this study, we screened a library consisting of 1,283 FDA-approved drugs and acquired by Ministry of Health of Mexico in order to identify potential inhibitors of SARS-CoV-2–ACE2 interaction.
Molecular modeling, electric partial-charge assignation, ligand conformer, searching of potential binding sites, energy minimizations, visualization and docking were performed with Molecular Operating Environment package26.
The chemical structure of 1,283 drugs that comprises the updated list of reference drugs, as well as the National Compendium of Health Supplies of Mexico (June 2020 update) were obtained from the DrugBank, ZINC15 and PubChem database in September 2020. In order to simulate ligand flexibility for our rigid docking simulations, we generated a set of low-energy conformer for each drug with Conformer Import tool, with an imposed conformational cut-off energy of 3 kcal/mol from minimum energy structure of each compound, calculated with the AMBER10-EHT force field. The resulted in-house molecular data base (mdb) contain multiple conformers for each molecule and were used for rigid docking simulation.
The X-ray crystal structure of SARS-CoV-2 RBDS1 in a complex with the ACE2 (PDB ID: 6VW1, resolution of 2.68 Å) was selected as a protein target for docking simulations. Importantly, this engineered structure is the first to presents all the functionally important epitopes in the SARS-CoV-2 receptor binding motif20. Potential binding sites in ACE2 near the interface region between the SARS-CoV-2 RBDS1 and ACE2 proteins, were identified with Site Finder tool. All crystallographic water and ligands molecules were removed from the system (chains B, E, F). Hydrogen atoms (Protonate 3D tool) and partial charges (Potential Setup tool) were added to ACE2 assuming pH equal to 7.0 and using the AMBER10-EHT force field, respectively. Before docking, the ACE2 protein structure was subjected to energy minimization using the same forcefield, in order to optimize atomic contacts. Docking simulations between the optimized ACE2 structure and each of the conformers contained in the in-house database, was carried out under the rigid-docking protocol. The docking parameters were set to take each ligand conformation as unique molecule, using the Alpha Triangle algorithm as placement method (at least 100 different orientations or poses on potential binding site) and further evaluation keeping the thirty best poses accordingly the London scoring function for binding affinity with a second refinement as a Rigid Receptor using Affinity dG algorithm keeping the ten best poses. The results were analyzed by docking score, frequency of the chemical compound as a stable conformation and the types of interactions at the binding site residues.
The structural analysis for the SARS-CoV-2 RBDS1 of the spike protein in a complex with the ACE2 (PDB ID: 6VW1; Figure 1A) revealing a potential site for ligand binding inside ACE2 structure (Table 1). The identified receptor site (Figure 1B) is proximal to the binding site of RBDS1 with a size of 86, therefore it can be used for simulating rigid molecular docking since receptor atoms are in an exposed region of the structure, which could be in favor of drug binding.
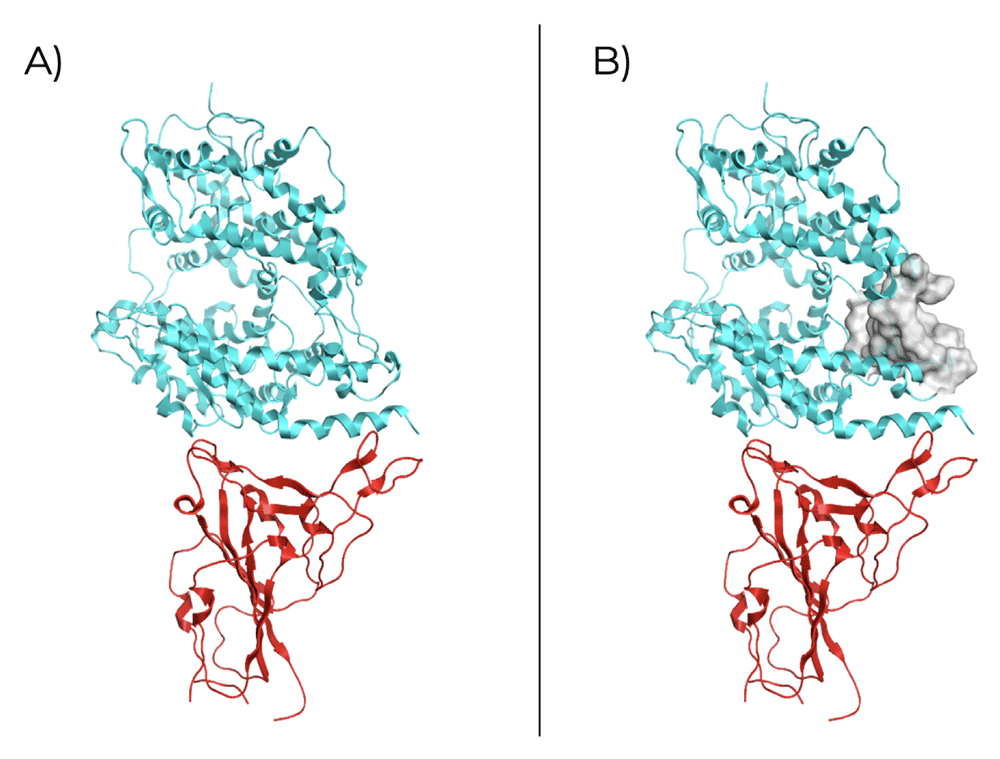
A) Crystallographic structure (PDB ID: 6VW1) of RBDS1(red) and ACE2 receptor (blue); B) Molecular surface of the selected binding site in ACE2.
An average of 78 conformations were generated for each ligand by Conformation Import MOE, generating 100,450 ligand conformations of the FDA approved and prescript drugs by the Mexican Public Health System.
The docking results were sorted and analyzed based on their S score, binding frequency which the drug binds to the receptor site and type of interactions, preferably, hydrogen bond, of the ligand with the selected site. We selected 38 drugs (Table 2) that presents the best docking score between −10.04 and −4.04.
Subsequently, we shortlisted nine drugs based on their risk of teratogenicity, route of administration, interaction with other drugs, side effects and by their background as pharmacological therapy for the treatment of respiratory diseases27,28.
Fexofenadine showed interactions of hydrogen bond with Lys 74, Ala 99, Ser 105, Ser 106, Trp 203 and Asp 509 (Figure 2A) with a docking score of −7.89. Pitavastatin displays hydrogen bond interaction with Gln 102, Tyr 196, Asp 206 and pi-H stacking with Leu 73 (Figure 2B) and a docking score of −8.40. Arformoterol showed hydrogen bond interactions with Tyr 202 and Asp 206 (Figure 2C) with a docking score of −6.85. Formoterol presented hydrogen bond interactions with Gln 98, Asn 194, ionic interaction with Glu 208 and pi-H interaction with Leu 85 (Figure 2D) and presents a docking score of −6.15. Ipratropium exhibited hydrogen bond interaction with Gln 98, Gln 208 and pi-H stacking with Asp 206 (Figure 2E) and a docking score of −5.38. Pargeverine shows hydrogen bond interactions with Gln 98, Gly 205, Glu 208 and ionic interaction with Glu 208 (Figure 2F) and presents a docking score of −4.53. Cholecalciferol presented hydrogen bond interaction with Gln 102 (Figure 2G) and had a docking score of −5.81. Lopinavir displays hydrogen bond interaction with Gln 102, Tyr 196 (Figure 2H) and a docking score of −8.62. Cefixime showed hydrogen bond interaction with Gln 98, Tyr 202, Glu 208, Arg 219, Lys 562 and ionic interaction with Arg 219, Lys 562 (Figure 2I) and a docking score of −9.02.
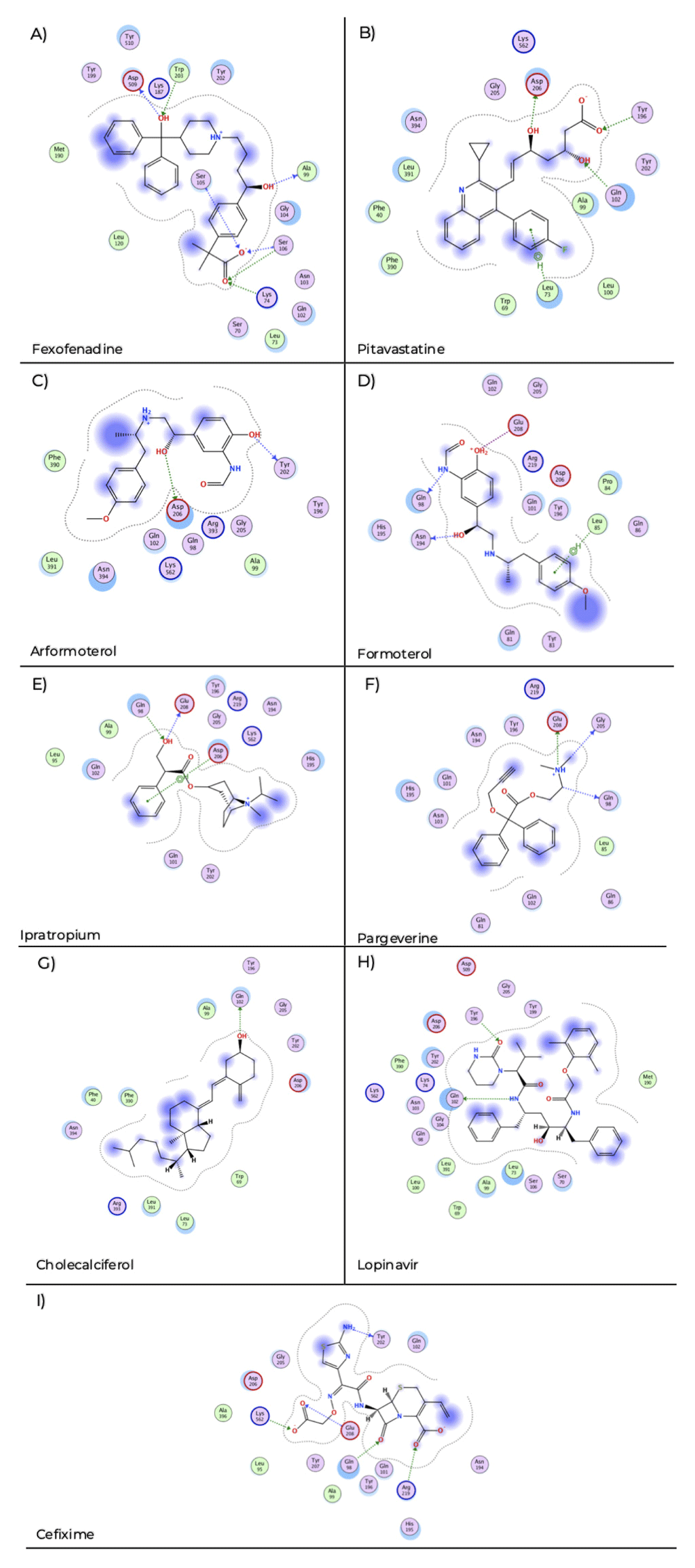
The blue arrows indicate the structural hydrogen bridge bonds, and the green arrows are the hydrogen bridge bonds with the side chain. A) Fexofenadine, B) pitavastatine, C) aformoterol, D) formoterol, E) ipatropium, F) pargeverine, G) cholecalciferol, H) lopinavir and I) cefixime.
Some pharmacokinetics characteristics27,28 of the shortlisted potential inhibitors of the RBDS1–ACE2 interaction are summarized in Table 3.
It has been established that S protein of SARS-CoV-2 virus plays a major role during viral infection. The S protein mediates receptor recognition, cell attachment and fusion of viral membrane with host cell membrane29–34. The S protein binds to ACE2 receptor through the RBDS1, mediating viral attachment to host cell35. Expression of ACE2 is ubiquitous in lung, intestine, heart and kidney, also alveolar epithelial type II cells had higher expression levels23. SARS-CoV-2, as one RNA viruses, has shown a high mutation rate as a result of lack of proofreading mechanisms, which leads to gain the ability to rapidly adapt to changes in their environment, which in turn leads to a great challenge for treating and preventing infections36. In this sense, the RBD region is a critical therapeutic target (vaccines and drugs) due to its indispensable function; however, it is suggested that mutations in this region may render pharmacological or immunological therapies ineffective37,38, therefore, it is needed to search and design alternative treatments.
In order to block this event, we propose an in silico approach to identify potential inhibitors of the SARS-CoV-2 –ACE2 interaction aiming at the ACE2 receptor, blocking the virus accessibility to the membrane-bound ACE2. In this sense, Jia and collaborators25 present an extensive review for this underexplored approach to treat COVID-19, pointing that it is imperative to determine, by clinicians, the stage of the disease and comorbidities that could prove consequential for an ACE2-targeting regimen. Here, we screened a drug library consisting of 1,283 drugs, FDA approved and prescribed by the Mexican Public Health System, for potential SARS-CoV-2−ACE2 inhibitors, using a rigid receptor docking approach. Utilization of an FDA-approved drug library is an effective and ideal tool for drug repurposing in antiviral research39,40, such as zika virus41, human rhinovirus42 and hepatitis B virus43. We identify 38 potentially inhibitor drugs of SARS-CoV-2−ACE2 interaction and these are listed on Table 2. Several of those drugs were previously reported to be used for the treatment of respiratory diseases.
Within this list of potential inhibitors of the SARS-CoV-2−ACE2 interaction, is fexofenadine, a third generation antihistamine whose therapeutic indication is the treatment of symptoms of stationary allergies through the selective blockade of H1 receptors44. It possesses direct effect on combating the cytokine storm caused by SARS-CoV-2 through inhibition of histamine and interleukin-6 (IL-6) release. In silico evidence45,46 suggest that it may interact with the SARS-CoV-2 main protease enzyme MPro, a key enzyme in viral replication47, acting as a potential inhibitor.
Cefixime is a third-generation antibiotic derived from cephalosporin whose use is indicated for the treatment of infections in the upper and lower respiratory tract, otorhinolaryngological48 and urinary tract49, inhibiting the synthesis of the bacterial wall by binding to specific binding proteins for penicillin and is currently used as a secondary therapy to prevent opportunistic infections during the development of COVID-1950.
Pitavastatin is a statin indicated for lowering blood cholesterol levels by inhibiting HMG-CoA reductase, preventing cholesterol synthesis51, and it has also been observed that statin treatments can interfere with viral infectivity through inhibition of glycoprotein processing52 also, they modulates the inflammatory process at cellular level53, which is a remarkable characteristic of the SARS-CoV-2 infection. Additionally, in silico findings suggest that could be an efficient inhibitor of SARS-CoV-2 MPro54 and SARS-CoV-2 RNA-dependent RNA polymerase (RdRp)55 thru active site binding.
Lopinavir is a protease inhibitor indicated as first barrier therapy, in conjunction with Ritonavir, to treat infection caused by the HIV virus by inhibiting the HIV-1 protease56 in addition, studies in cell cultures have shown its effectiveness as an inhibitor of the replication of the MERS-CoV virus57 and SARS-CoV-158, while in severe cases of SARS-CoV-2 infection, the results of clinical trials indicate that it is not useful59.
Formoterol and aformoterol, anenantiomer of formoterol, are long-lasting selective β agonists indicated for the treatment of chronic obstructive pulmonary disease (COPD) and bronchospasms60, in the same way there is evidence of the use of these drugs as a partial inhibitor of viral replication in primary epithelial cells cultures61 and in silico data suggest their binding to the papain-like protease PLpro, a coronavirus enzyme essential for viral spread62.
Ipratropium is a bronchodilator anticholinergic indicated for the treatment of asthma, shortness of breath, cough and tightness in the chest in patients with COPD63,64. Inhalation therapy with ipratropium is currently in use to dilate bronchioles in COVID-19 patients to increase oxygen saturation levels (from <80% to 94%)65.
Pargeverine is an antispasmodic opioid alkaloid whose therapeutic indication is aimed at the treatment of painful spasms66, also, acts as anticholinergic and has a moderate and non-selective blockade of muscarinic cholinergic fibers67. Since cholinergic activity contribute to airway narrowing, this might be a potential agent to open airway obstruction.
Cholecalciferol, is a form of vitamin D (vitamin D3) that can be synthesized naturally in the skin and acts as a hormonal precursor, being converted into calcitriol, and therapeutically is used as a vitamin supplement to treat deficiencies of this vitamin68. In addition, it has been observed that vitamin D supplementation is favorable to reduce viral infections such as influenza69,70 or more aggressive cases such as HIV71 and it has recently been suggested that it also presents favorable effects before and during the infection caused by SARS-CoV-272.
Likewise, it is important to take into account that ACE2 plays an important biological role since regulates cardiovascular functions and innate immune system73 and, therefore cautions must be taken. Another point to consider is the delivery method of the drug, since the primary target must be smooth muscle, like the one surrounding the bronchioles, and lung epithelial cells in the airway and airspace compartments, hence, inhalable delivery would be the acceptable choice to deliver the drug in a selectively and localized manner.
Given these characteristics, the results obtained through our in silico approach, we consider that the aforementioned drugs are outlined as possible inhibitors of the RBDS1-ACE2 interaction. These drugs are well tolerated, commonly used and affordable, hence, most of the drugs on this list can be tested in vitro, and even in vivo and, consequently, in clinical trials for the development of adjuvant therapies to treat COVID-19.
In the absence of approved therapies for treatment or prevention, drug repurposing has provided fast and valuable insight into the treatment of COVID-19. Targeting ACE2 receptor as a COVID-19 therapy it is a conceivable approach since it is essential for the viral internalization. However, this approach requires an integrative evaluation of the pros and cons by a clinical context since ACE2 is a multifunctional protein. Several drugs are currently investigated by clinical trials or are already in use to treat COVID-19 patients, like lopinavir or ipratropium. In this in silico study using structure-bases virtual screening, we identified potential inhibitors of SARS-CoV-2–ACE2 by their interaction with ACE2 receptor. Based on desired characteristics like pharmacokinetics, route of administration or by their background as pharmacological therapy, we propose a shortlist of drugs suitable for testing their potential RBDS1 –ACE2 inhibitory activity: fexofenadine, cefixime, pitavastatine, lopinavir, arformoterol, formoterol, ipratropium, pargeverine and cholecalciferol. Our identification of potential inhibitors of the SARS-CoV-2–ACE2 interaction among commonly use drugs highlights their potential use for treating COVID-19. Further in vitro, in vivo or clinical trial are needed to validate their potential use as inhibitors of SARS-CoV-2–ACE2 interaction.
Protein Data Bank: Crystal structure of SARS-CoV-2 receptor binding domain (RBDS1) of the spike protein in a complex with the ACE2 receptor. https://identifiers.org/pdb:6vw1.
Ministry of Health of Mexico: Drug list used by the Ministry of Health of Mexico (June 2020 version). http://www.csg.gob.mx/Compendio/CNIS/cnis.html.
PubChem: Ligands. https://pubchem.ncbi.nlm.nih.gov74.
Drugbank: Ligands. https://go.drugbank.com75.
ZINC database: Ligands. http://zinc.docking.org76.
Figshare: Identification of potential inhibitors of SARS-CoV-2 S protein–ACE2 interaction by in silico drug repurposing, https://doi.org/10.6084/m9.figshare.1446633377
This project contains the underlying data file:
Data is available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0)
FET-F and DC-A performed most of the bioinformatics work, shared first authorship, equal contribution to this work; FET-F wrote the initial manuscript draft; JAC-M and RP-A generated the ligand data base and assisted bioinformatic analysis; GA-G revising critically the initial manuscript draft and provision facility resources; P.G.-G, DC-A and GAS-M contributed intellectually to the project conceptualization and participate in the initial manuscript draft; DC-A and G.A.S.-M designed and supervised the project. G.A.S.-M conceptualize, acquire the financial support for the project leading to this publication, supervised the bioinformatic work and wrote the final version of the manuscript; all authors read and approved the final manuscript version.
The authors would like to thank Ruy E. Pineda-Silva for the supporting work in data acquisition; Martha C. Silva-Martinez, MD. and Omar Pineda-Gama, MD. for advice, assistance, and their valuable feedback.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Medical devices development, Medical Imaging, Methods development
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Pharmacy and Pharmacology
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Pharmacy and Pharmacology
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Medical devices development, Medical Imaging, Methods development
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 11 Nov 21 |
read | read |
|
Version 1 07 May 21 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)