Keywords
K. Pneumoniae, ESBL, TEM, SHV, CTX-M-15
K. Pneumoniae, ESBL, TEM, SHV, CTX-M-15
In response to reviewers comments the following updates were done.
Updated Title: Molecular Epidemiological Surveillance of CTX-M-15-producing Klebsiella pneumoniae from the patients of a teaching hospital in Sindh, Pakistan.
Reason for update: As suggested by reviewer
Abstract:
Updated the text: This study was aimed at the molecular characterization of Extended-spectrum β-lactamase -positive Klebsiella pneumoniae isolates recovered from the patients of a teaching hospital in Sindh, Pakistan.
Reason for update: Clearfield the objective.
Discussion:
Updated text: Larger studies need to be done in various geographical regions of the country to better define the molecular epidemiology of ESBL-producing K. pneumoniae and its clinical implications.
Reason for update: Provided details about limitations of the study.
References:
Updated text: Naeem, S., Bilal, H., Muhammad, H., Khan, M. A., Hameed, F., Bahadur, S., & Rehman, T. U. (2021). Detection of blaNDM-1 gene in ESBL producing Escherichia coli and Klebsiella pneumoniae isolated from urine samples. Journal of infection in developing countries, 15(4), 516–522. https://doi.org/10.3855/jidc.12850
Reason for update: Included recent relevant reference added.
See the authors' detailed response to the review by Abbas Farahani
See the authors' detailed response to the review by Mubashir Zafar
See the authors' detailed response to the review by SUPRIT DESHPANDE
The emergence of Extended-spectrum β-lactamase (ESBLs) producing organisms has imposed a great threat on majority of the antibiotic classes, particularly cephalosporins.1 The situation worsens when the patient infected with ESBL-producing organism is administered an antibiotic to which the organism is resistant.1 Clinical practitioners globally have been facing problems related to plasmid mediated ESBL producing Klebsiella pneumoniae (K. pneumoinae). These plasmids contain genes encoding resistance to antibiotics such as aminoglycosides, sulfonamides, tetracycline, chloramphenicol, and quinolones.2 Cephalosporins are β-lactam antibiotics that are widely used in clinical practice and in the treatment of bacterial infections.3 In addition, K. pneumoniae is reported in nosocomial infections and community acquired infections.4
A study conducted by Maltezou et al.5 revealed that the incidence rate of ESBL producing K. pneumoniae infection was 16.71% for Northern Europe, 24.41% for Southern Europe, 58.71% for Eastern Europe, 28.2% for the Asia Pacific region, 51.9% for South America, and 12.3% for North America.5
CTX-M-type β-lactamase, has become more predominant than conventional SHV and TEM-type ESBL, which have covered an extensive range of clinically significant bacteria and over a wide environmental area.6 Moreover, strains that yield ESBL often reveal resistance to antibiotics belonging to other classes (i.e. aminoglycosides, quinolones, and sulfonamides); making its management more complicated.7
Consequently, this class of bacteria makes a need to ensure the reporting of ESBL producers in clinical isolates and detection of newly emerging drug resistant isolates. The changing trends of drug resistant ESBL-producing K. pneumoniae need time to watch for management, confinement and isolation of patients in hospitals before discharge. It is also necessary to monitor the prevalence and types of ESBLs and define the appropriate therapeutic options accordingly. Hence, the purpose of the current study was to observe the current trends of ESBL producing K. pneumoniae drug resistant patterns and to identify the occurrence of ESBL genes in Sindh region of Pakistan.
A total of 1458 non-duplicate clinical samples were collected between June 2019 and May 2020 from outpatient and inpatient wards of People's University of Medical and Health Sciences, Nawabshah, Pakistan [tertiary care government hospital with 1000 beds] through its diagnostics and research laboratory. Patients referred by their primary physicians for screening tests (such as blood complete count and erythrocyte sedimentation rate (ESR) were recruited in the study. The clinical samples including blood, sputum, urine, cerebrospinal fluid, tracheal secretions, and pus samples were collected by standard techniques (Figure 1A). The clinical samples, which yielded the K. pneumoniae were selected for further assessment, while others were excluded. The demographic data was retrieved from clinical survey that included age, gender, specimen type, town, and susceptibility patterns (Table 2).
The samples were not specifically collected for this research, they were used after the completion of routine laboratory investigations upon the permission of the concerned laboratory's authority. The isolation of bacteria was carried out by conventional bacteriological techniques and identification was confirmed by API 20E identification kit (BioMerieux, France).8 As many as 513 K. pneumoniae isolates from various clinical samples and sites (Figure 1A) were obtained and included in the present study.
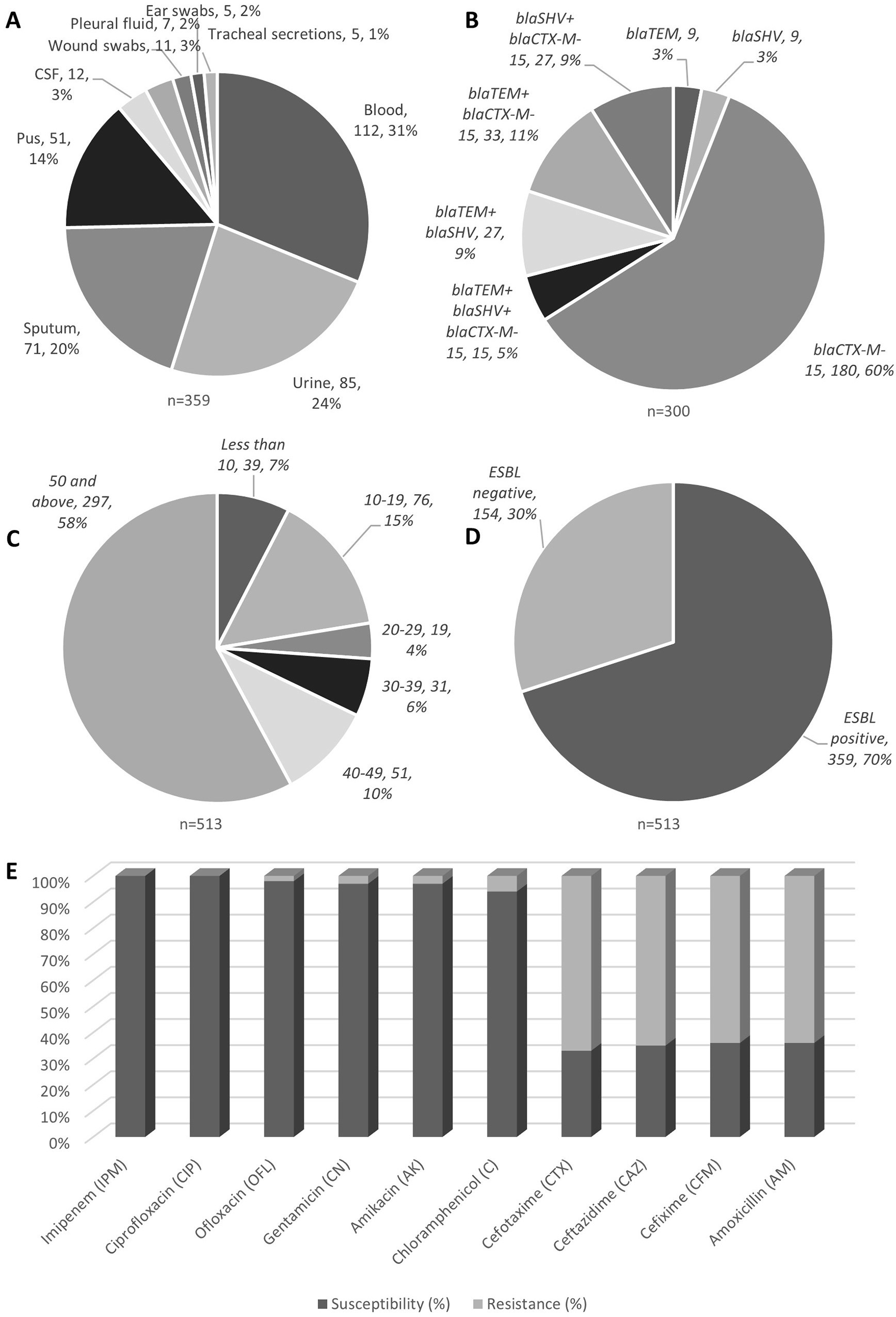
K. pneumoniae isoaltes obtained from various clinical samples showed high prevalence as blood borne infections (A); genotypic distribution showed blaCTX-M-15 to be highly prevalent (B); age group wise analysis showed elderly group of 50 years old and above showed high incidences (C); ESBL positivitiy to be 70% among all isolates tested (D), with their antibiotic susceptibility profile (E).
Antimicrobial susceptibility testing (AST) was performed by Kirby Bauer Disk diffusion method on Mueller-Hinton agar plates (Beckton Dickinson, Sparks, MD, USA) according to Clinical Laboratory Standards Institute (CLSI).8 Disks (Beckton Dickinson) comprising of the following antibiotics were used: cefixime 30 μg, amoxicillin 30 μg, ceftazidime 30 μg, cefotaxime 30 μg, Ofloxacin 10 μg, imipenem 10 μg, amikacin 30 μg, gentamicin 10 μg, ciprofloxacin 5 μg and chloramphenicol 30 μg.
Double disk diffusion methods were used to detect ESBL using four antibiotic disks: cefotaxime 30 μg, cefotaxime with clavulanic acid 10 μg, ceftazidime 30 μg, and ceftazidime with clavulanic acid 10 μg. A more than 5 mm increase in zone of inhibition width for whichever antimicrobial agent tested in combination with clavulanic acid against its zone of inhibition when tested alone was labelled as ESBL positive.9 The inoculum and incubation conditions were the same as recommended for standard disk diffusion method. K. pneumoniae ATCC 700603 (American type culture collection, ATCC, Manassas, USA) was used as quality control strain.
Bacterial genomic DNA was seperated by boiling the young bacterial colony in 50μl distilled water in a hot bath set at 98°C. This DNA template was used for all PCR reactions. Amplification of blaTEM, blaSHV and blaCTX-M-15 ESBL genes was done on GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA). The primers used are given in Table 1. The PCR yields were run on 1% agarose gels and observed in UV light. Purified PCR products of CTX-M genes were sequenced on an Applied Biosystems 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA).
| ESBL Gene | Primer | Sequence (5′-3′)* | Product size (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| blaTEM | Forward | ATGAGTATTCAACATTTCCGTG | 861 | 55 |
| Reverse | TTACCAATGCTTAATCAGTGAG | |||
| blaSHV | Forward | ATTTGTCGCTTCTTTACTCGC | 1051 | 60 |
| Reverse | TTTATGGCGTTACCTTTGACC | |||
| blaCTX-M-15 | Forward | CACACGTGGAATTTAGGGACT | 996 | 50 |
| Reverse | GCCGTCTAAGGCGATÁAACA |
Reaction mixture: A 30 μl total reaction mixture consisted of the following: 2 μl of DNA template; 1.5 μl of 10 μM forward and reverse primer; 1.5 μl of Sigma brand Red Taq; 20.2 μl of pure water; 3 μl of PCR buffer with MgCl2; and 0.3 μl of 10 mM dNTPs.
The data were analyzed by IBM SPSS Statistics for Windows, Version 21.0, Inc. USA Statistical Package of Social Sciences (SPSS) version 21.0. Continuous and categorical variables were analyzed by t-test and Chi-square testing, respectively (these tests can also be carried out on the open source software R). Variance of resistance levels of various antimicrobials in ESBL producing isolates versus non-ESBL producing isolates were calculated by the Fisher exact test. The data was analyzed at 95% Confidence interval (p ≤ 0.05 indicates a significant difference).
During the study period, a total of 513 (35.18%) K. pneumoniae were isolated. From those 513 positive cultures, 411 (80.1%) belonged to males and 102 (19.8%) to females (p = 0.0001). The demographic characteristics of patients including age, gender, rural or urban, and clinical interventions are summarized in Table 2. The results show that the majority of patients were in the older age group (50 and above) and lived in urban areas Figure 1B. The categories of patient's ages have been shown in Figure 1C. The mean age of the total study subjects was 48.5 ± 8.9 years.
Out of 513 isolates 359 (69.9%) were ESBL producing K. pneumoniae (Figure 1D). The incidences of ESBL was higher in males as compared to females. The frequency of ESBL producing K. pneumoniae from different samples (n = 359) has been shown in Table 3 and Figure 1A. The means ± SD age of patients with ESBL positive and negative K. pneumoniae was 47 ± 17.5 years and 47.5 ± 12.8 years, respectively (p = 0.093).
The results show that 64% of K. pneumoniae in our sample were resistant to Amoxicillin and Cefixime. In addition, an important drop of 30–40% was perceived in the susceptibility for all Cephalosporins. However, K. pneumoniae presented a dissimilar sensitivity rate to Chloramphenicol, Ofloxacin, Gentamicin and Amikacin with 94%, 98%, 97%, and 97%, respectively and depicted in Table 4.
Regarding the PCR and DNA sequencing of ESBL genotypes, it was found that all of the ESBL-positive K. pneumoniae isolates had one or more ESBL genes that were tested in the present study and presented in Table 5. Overall, 83.56% (300/359) of K. pneumoniae isolates were positive for one or more ESBL genes. The PCR assay and DNA sequencing results indicated the following frequencies of ESBL genotypes: 85% blaCTX-M-15 gene, 26% blaSHV gene, and 28% blaTEM gene.
| Genotypes | Incidences n (%)* |
|---|---|
| blaTEM | 9 (3%) |
| blaSHV | 9 (3%) |
| blaCTX-M-15 | 180 (60%) |
| blaTEM+ blaSHV+ blaCTX-M-15 | 15 (5%) |
| blaTEM+ blaSHV | 27 (9%) |
| blaTEM+ blaCTX-M-15 | 33 (11%) |
| blaSHV+ blaCTX-M-15 | 27 (9%) |
ESBL's are the major causes of β-lactam antibiotic resistance, particularly in K. pneumoniae, which is amongst the most common gram-negative bacteria belonging to Enterobacteriaceae families.10 A study from Ziauddin University Karachi, Pakistan has showed that ESBL producing K. pneumoniae frequency is 84.16% amongst reported cases.11 Similarly, Saleem et al. (Aga Khan University, Karachi, Pakistan) have reported a frequency of 80% of ESBL producing K. pneumoniae.12 These findings were higher as compared to the results presented in the current study, which showed that the frequency of ESBL producing K. pneumonia is 69.6% with a higher incidence in male than in female patients. In this repect, the frequency of EBSL producing K. pneumoniae varies from one institution/hospital to another. Several factors could govern this variation such as; the use of antibiotics, drug dose, infection control measures, treating physicians, and the duration of drug therapy - all of these elements may cause a difference in results. The frequency of ESBL producing K. pneumoniae in previous research conducted at Aga Khan University Karachi, Pakistan was 30.1% and 47.8%, which is low compared to the present study, as well as, previous studies conducted by Mansouri et al.10 and Ejaz et al.13 A Pakistani study found a frequency of ESBLs producing K. pneumoniae of 70%,14 which is in agreement with our study.
Carbapenems are often the final influential therapy of choice to treat infections resulting from MDR Enterobacteriaceae. Based on our study and other studies, 100% sensitivity was seen with Imipenem, which have been found to be the most effective antibiotics on the isolates that produce ESBLs. This is an important result of the present study because many infections can be treated with Carbapenems.12,15
blaCTX−M is a predominant genotype in this area of the subcontinent. A further study from Pakistan has reported that 72% of ESBL producing strains had the blaCTXM−15 gene, which was higher than the prevalence of blaCTX-M gene reported in the present study.16 Limited studies from different regions have also shown a higher prevalence of the blaCTX-M genotype amongst strains including 84.7% (Chile), whereas a very high prevalence of 98.8% was reported in China and, significantly, a low prevalence of 13.6% in Tanzania.17–19
In conclusion, the findings of the current study emphasizes the urgent need for screening and surveillance of ESBL producing K. pneumoniae in routine microbiology laboratories for early detection, prompt intervention, and successful clinical management before exacerbation of the infection magnitude. In this regard, the current findings may be a valuable contribution to the medical literature providing physicians with an updated prevelance status of ESBL producing K. pneumoniae. Larger studies need to be done in various geographical regions of the country to better define the molecular epidemiology of ESBL-producing K. pneumoniae and its clinical implications.20 Finally, the limited sample size along with the limited time frame are two common limitations, which could affect the outcomes of the study.
The authors greatly acknowledges the assistance received from Dr. Omar Soliman Mohamed Elmasry, Imam Abdulrahman Bin Faisal University, Dammam, through various stages of this study. The authors are very thankful to the Diagnostic and Research Laboratory, People's University of Medical and Health Sciences (PUMHS) Nawabshah, Sindh, Pakistan, for their contribution to this research.
Prior to the study we contacted the institutional review board representative (PUMHS, IRB) for discussion. The IRB representative confirmed that since patients are not directly involved and the samples are not collected specifically for this research there is no need for IRB approval. Patient data consent was not required as we didn't retrieve any personal information, such as medical record numbers, national ID, and names, from patients. The data was completely anonymous and thus does not violate the privacy or confidentiality of any of the patients.
All data underlying the results are available as part of the article and no additional source data are required.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Microbiology
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
No
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Preventive Medicine specialist
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: HIV molecular virology , immunology and antibody discovery
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Microbiology
Is the work clearly and accurately presented and does it cite the current literature?
No
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
No
Are all the source data underlying the results available to ensure full reproducibility?
No
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Microbiology
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: HIV molecular virology , immunology and antibody discovery
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 3 (revision) 15 Nov 21 |
read | read | |
|
Version 2 (revision) 23 Jul 21 |
read | read | read |
|
Version 1 04 Jun 21 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)