Keywords
disease activity, HLA-B*27, HLA-C, HLA-K, HLA Class I, Indonesia, spondyloarthritis
This article is included in the Genomics and Genetics gateway.
This article is included in the Cell & Molecular Biology gateway.
disease activity, HLA-B*27, HLA-C, HLA-K, HLA Class I, Indonesia, spondyloarthritis
Spondyloarthritis (SpA) is a chronic inflammatory disease characterized by enthesitis, sacroiliitis, and axial joint involvement. SpA is a group of several linked diseases but phenotypically distinct disorders that consist of psoriatic arthritis, reactive arthritis, enteropathic arthritis, and ankylosing spondylitis.1 The classification and diagnosis of SpA are by depicting the sacroiliac joint, hip joint, enthesitis, or the presence of human leukocyte antigen B27 (HLA-B*27).2 Inflammatory changes in the sacroiliac joint can be detected by magnetic resonance imaging (MRI) in the early stages of SpA in patients, but there is usually a five to 10-year delay from onset to the diagnosis of SpA.
Genetic identification of HLA-B*27 has good specificity and sensitivity, and it is also relatively inexpensive compared to MRI for the diagnosis of SpA, but the positive rate of HLA-B*27 in healthy individuals is relatively high, up to 10%.1,3 In addition to HLA-B*27, there is an association of other genes in HLA class I with SpA, including various subtypes of HLA-A and HLA-C, but despite this the level of this correlation has not been clearly reported.4–6 Determining the HLA alleles involved in the disease activity of SpA is important because different alleles affect the 3D structure, biochemical activity, and peptide-binding groove of HLA proteins.7–9
Based on the clinical experience of 15 years of observations, there are an increasing number of SpA patients and severe clinical manifestations in the outpatient clinic at Dr. Soetomo Hospital, Indonesia. There are also many patients with poor clinical manifestations of early disease. The number of SpA patients increases with a growing population, genetic changes, environmental factors, unhealthy lifestyles, and other random events.10,11 Defective DNA repair can lead to DNA damage, mutations, and cellular dysfunction, typically associated with many diseases, including autoimmunity.12 Genetic mutations can also generate additional non-coding regions in the human genome, such as the emergence of pseudogenes. Pseudogenes resemble functional genes due to similarity with the parent gene despite having coding-sequence deficiencies like frameshift mutations and premature stop codons.13
The high prevalence of SpA and correlation with the HLA class I alleles make research on the subtypes that correlate with SpA in Indonesian society crucial. Nevertheless, an updated report is needed as the last report on HLA alleles in Indonesian samples was carried out in 1997.14 The recognition of appropriate HLA alleles can provide clear evidence for the diagnosis of SpA. This study aims to identify the HLA class I alleles in SpA patients at Dr. Soetomo General Hospital in order to extend understanding of HLA class I alleles related to the pathogenesis of SpA, especially in specific ethnic groups such as Indonesia.
This study used a cross-sectional analytical design with 70 subjects that met the criteria for spondyloarthritis according to the Assessment of SpondyloArthritis international Society (ASAS) 2009 at the Dr. Soetomo Hospital outpatient clinic. These patients were clinically evaluated for their history of disease and had a clinical examination. Clinical scorings with modified Stoke Ankylosing Spondylitis Spinal Score (mSASSS), Bath Ankylosing Spondylitis Functional Index (BASFI) score, Ankylosing Spondylitis Disease Activity Score (ASDAS), and Schober test were used to evaluate the disease activity. The patient's blood collection was carried out at Dr. Soetomo General Hospital, then the serum preparation process, DNA extraction, polymerase chain reaction (PCR), and sequencing were carried out at the Institute of Tropical Disease, Airlangga University. The health research ethics committee at Dr. Soetomo General Hospital approved this study (0203/KEPK/V/2021). Before participating in this study, the patients signed a written informed consent in which confidentiality of all information was guaranteed. This study was also conducted in accordance with the principles contained in the Declaration of Helsinki for studies on humans.
The HLA class I genes were genetically analyzed using PCR, and further Sanger sequencing was performed to determine the nucleotide sequence to identify the HLA types. For each blood sample, 200 μL serum was extracted using the AuPreP GEN DNA extraction kit (Life Technologies India Pvt Ltd., New Delhi, India) according to the manufacturer's protocol. The PCR mixture of each reaction was 25 μL of DreamTaq™ Green PCR Master Mix (Thermo Scientific™), 300 nM of forward primers, 300 nM of reverse primers, 5 μL of DNA template, and ended by adding the PCR grade water up to 50 μL of total volume. Primer designing for HLA Class I was carried out in this study using Lank et al.’s15 method with the NCBI database. Common conserved primer binding sites to HLA-A, B, and C alleles at 344–373 bp (5′) and 592-621 (3′) were used to design a 261 bp amplicon, and the sequences of the primers were; Ex3-F: 5′ACCCTCCAGAATATGTATGGCTG3′, Ex3-R1: 5′CTTCCCGTTCTCCAGGTATC3′. The amplification was performed at 40 cycles of 94°C for 30 seconds for denaturation, followed by annealing at 59°C for 30 seconds, and finished by the synthesis process at 72°C for one minute. The 261 bp of PCR product was then sequenced with an ABI Prism 310 Genetic Analyzer (Perkin Elmer).
After the HLA Class I sequence from the patient was obtained, the sequence was then aligned with the NCBI database using NCBI BLAST (RRID: SCR_004870) to determine the HLA types. A maximum-likelihood phylogenetic tree was then constructed with the Tamura 3-parameter with discrete Gamma distribution (+G) using MEGA 11 software (RRID: SCR_000667),16,17 and edited with iTOL v6 (RRID: SCR_018174). The HLA-B*27 (X03945.1) reference sequence from NCBI is also used in this tree. The pairwise comparison and phylogenetic tree analysis reliability was assessed by 1000-replicate bootstrapping.
Descriptive analyses and frequency distributions were performed for all clinical data. Interpretations using median with 95% CI were used for clinical scoring (mSASSS, BASFI, ASDAS, Schober) and duration of illness. A comparative analysis of clinical scoring and duration of illness by HLA type was performed using Mann–Whitney U test with p-values of <0.05 considered statistically significant. All analyses and graphic presentations were performed using GraphPad Prism v9 (RRID: SCR_002798) (An open-access alternative is JASP 0.16.3 for Windows (RRID: SCR_015823)).
Table 1 shows the characteristics of SpA patients at Dr. Soetomo General Hospital. In this study, there were 70 patients consisting of a 4:3 ratio of male and female, with the majority between 36 and 55 years old, and most of the patients had a normal BMI. Several symptoms experienced by more than half of the patients were back pain, psoriasis, and sacroiliitis with grade 4 bilateral. Some patients also experienced gastrointestinal disorders and fused spines (bamboo spine).
All samples in this study were positive towards the targeted band (~261 bp), as shown in Figure 1, these 70 samples were then followed up by sequencing for further examination to determine the type of HLA. The sequencing results (Table 2) showed the three types of HLA: HLA-B, -C, and -K, with the HLA-K being the most detected type in more than half of the total patients. There were seven alleles of HLA-B detected: HLA-B*2704, -B*2705, -B*2706, -B*1802, -B*57v, -B*1802, and -B53.
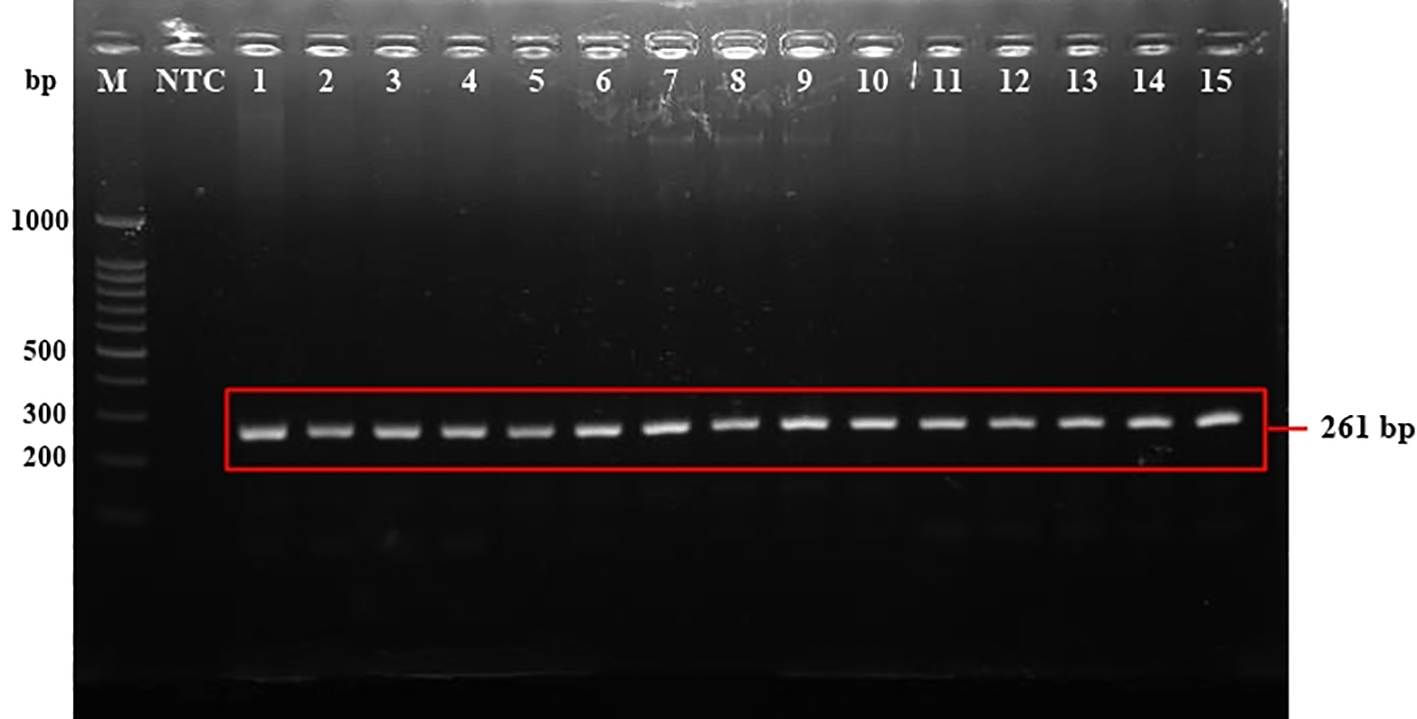
M: 100 bp DNA ladder. NTC: No template control. Lanes 1–15: Samples (Patients of Dr. Soetomo General Hospital). The electrophoresis is only represented by samples 1–15 due to the similarity of results in all samples.
| HLA type | n (%) | Highest similarity (%) | Accession number |
|---|---|---|---|
| HLA - B | |||
| -B*2704 | 9 (12.86) | 100 | GQ118997.1 |
| -B*2705 | 1 (1.43) | 100 | EF215529.1 |
| -B*2706 | 1 (1.43) | 98.53 | AJ550736.1 |
| -B*1802 | 3 (4.28) | 100 | GQ118994.2 |
| -B*57v | 1 (1.43) | 83.36 | AM889027.1 |
| -B*35 | 2 (2.86) | 100 | OM210002.1 |
| HLA - C | 15 (21.43) | 100 | MT108849.1 |
| HLA - K | 37 (52.83) | 100 | NG_015982.3no |
A phylogenetic tree was constructed to show the evolutionary relationships among samples. Figure 2 shows the branching that divided samples into two large groups: the HLA-B and HLA-K. The HLA-C is closely related to HLA-B, indicated by the spread of HLA-C along with the HLA-B group. The HLA-B*27 unified in clade II with two of the most frequently discussed HLA alleles associated with SpA, HLA-B*2704 and -B*2705 (* and **), are in this clade with HLA-B*2706. The other HLA-B subtypes clustered in clade I with HLA-B*35 and -57 were in one subclade, and HLA-B*1802 clustered in another. The HLA-K clustered in clade III is the most homogeneous subtype compared to other subtypes. Despite one HLA-K sample being separated, the sample did not form any group with other subtypes of HLA. Branch lengths indicate genetic changes, with YUL61 (HLA-K) having the longest branch on the phylogenetic tree indicating that this sample has the most genetic changes.
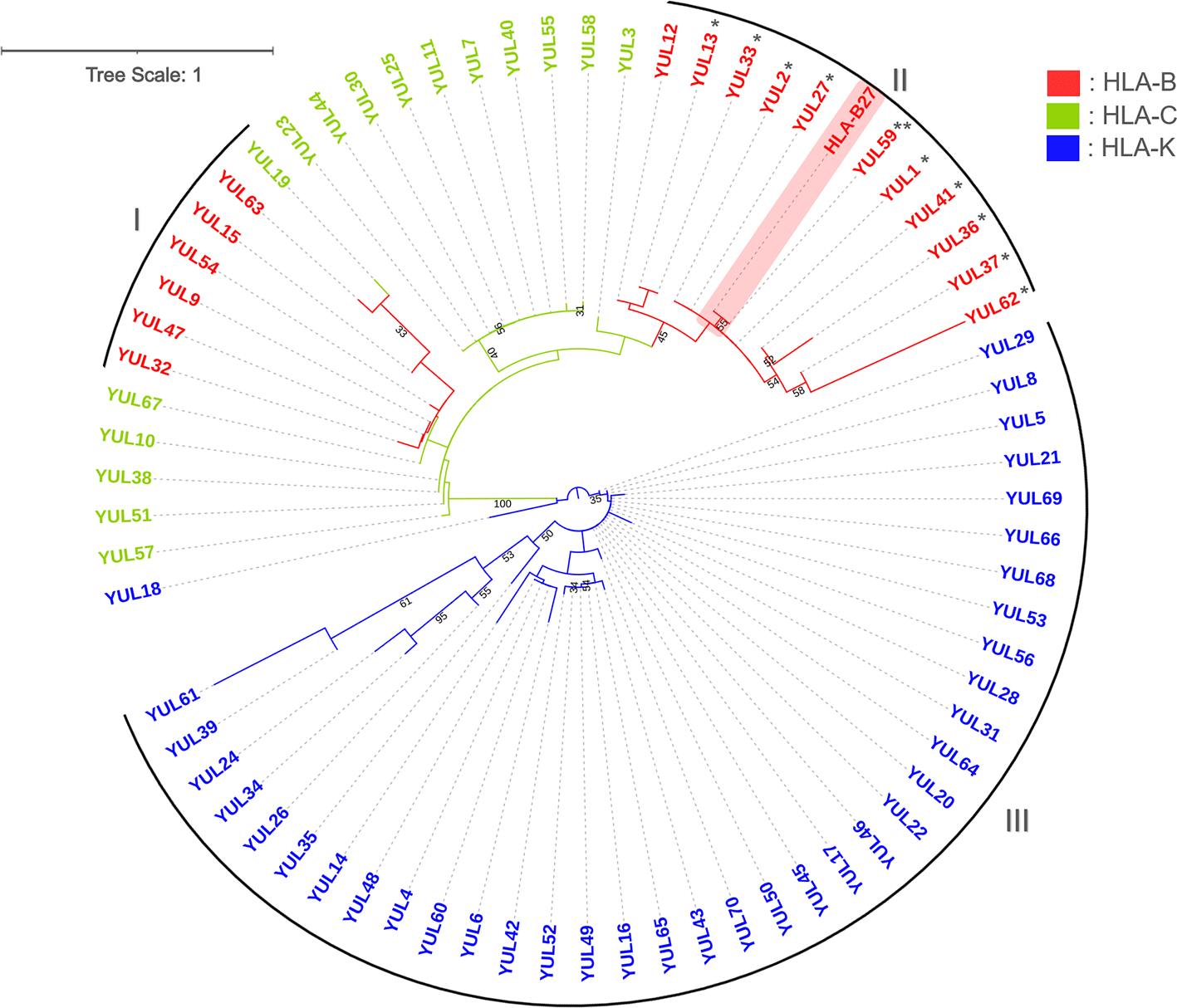
Constructed with Tamura 3-parameter with discrete Gamma distribution (+G). Numbers below branches indicate bootstrap (BS>30) support values. The HLA-B*27 (highlighted with red) from the NCBI database was used as the reference sequence. *: HLA-B*2704, **: HLA-B*2705. I-III: number of clades.
Figure 3 shows disease activity in each type of HLA, the median value was used to interpret this data due to the asymmetrical distribution. Median values of the mSASSS score of HLA-B*2705 and -B*2704 and the other HLA-B alleles were 20.5 (95% CI, 12–28) and 24 (95% CI, 18–54), while the medians of HLA-C and HLA-K were 24 (95% CI, 20–36) and 26 (95% CI, 22–28). A higher mSASSS score indicates poor severity, and the HLA-K has the worst severity compared to other types of HLA. In line with the mSASSS score, worse disease severity based on the BASFI score was also experienced by the HLA-C and HLA-K types with median values of 71 (95% CI, 59–97) and 60 (95% CI, 55–68), while the medians of BASFI scores for HLA-B*2705 and -B*2704 and other HLA-B were 59.5 (95% CI, 48–80) and 45 (95% CI, 35–75). The median of ASDAS values for all HLA types were: 4.46 (95% CI, 3.54–4.67), 3.94 (95% CI, 2.05–4.97), 4.18 (95% CI, 4.00–4.63), and 4.20 (95% CI, 3.80–4.25) for HLA-B*2705 and -B*2704, other HLA-B, HLA-C, and HLA-K, respectively.
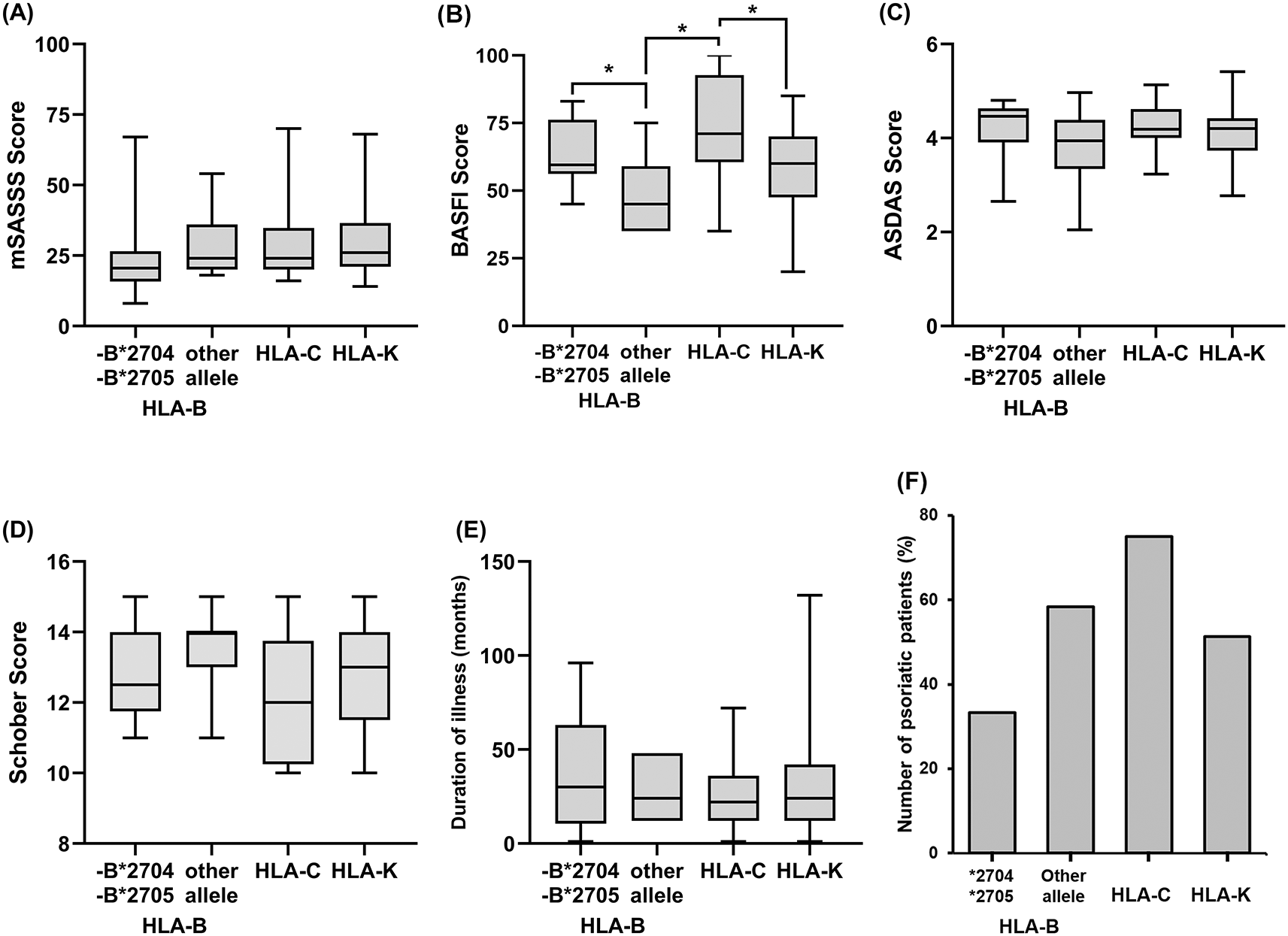
A: The mSASS score, B: The BASFI score, C: The ASDAS score, D: The Schober score, E: Duration of illness, F: Number of psoriatic arthritis. The percentage of psoriatic patients were calculated by each type of HLA. Bars on each boxplot graph indicate median and interquartile range (IQR). The p-values of <0.05 were considered statistically significant (*).
A low Schober score indicates a poor prognosis, and as with the other scoring, HLA-C and HLA-K show a lower Schober score with medians of 12 (95% CI, 10–14) and 13 (95% CI, 12–14) compared to the HLA-B*2705 and -B*2704 and other HLA-B with medians of 12.5 (95% CI, 11–14) and 14 (95% CI, 11–15). Patients with HLA-C had the shortest duration of illness with a median of 22 months (95% CI, 12–36), then HLA-K and other HLA-B alleles with a median of 24 months (95% CI, 24–36 and 12–48), and the longest duration of illness was HLA-B*2705 and -B*2704 group with a median of 30 months (95% CI, 6–72). The grouping of psoriatic patients based on HLA types showed that HLA-C had the highest percentage of 75%, while HLA-B*2705 and -B*2704 and HLA-C were 33%, 58%, and 51%, respectively. From the results mentioned earlier, the HLA-B*2704 and -B*2705 had less severe clinical symptoms than patients with HLA-C and HLA-K.
Identifying HLA types is crucial for SpA patients to provide a proper diagnosis and treatment. This study carried out genetic identification of the HLA types found in SpA patients at Dr. Soetomo General Hospital in Indonesia. Interestingly, not only HLA-B*27 was found. The HLA-B*2704 and -B*2705 subtypes, the most widely discussed subtypes associated with SpA, were detected in only 10 out of 70 patients. The HLA-B*1802, -B*35, B-57, and B-2706 subtypes were also found in patients, but they accounted for less than 5% of each type. Several studies suggest that HLA-B*2706 is a protective factor against SpA,18,19 but a study by Sudarsono et al.20 stated that HLA-B*2706 did not have any protective properties against SpA, especially in Indonesia. There are few studies on HLA-B*2706, primarily due to the fact that this subtype is rare.21 The role of other types of HLA-B against SpA disease found in this study is unclear.
We found that HLA-K was the most common subtype in SpA patients. From the NCBI database, the patient sequences that were detected as HLA-K were identical to the database with accession number NG_015982.3. This database also contains information that HLA-K is a pseudogene. Pseudogenes derived from protein-coding genes have lost their protein-coding capacity due to deleterious disruptions (e.g., premature stop codons or frameshift mutations) in their hypothetical open reading frames.22 The pseudogenes will precisely reflect the underlying mutation process. Therefore, pseudogenes can infer mutation processes for nucleotide changes in species and the effects of DNA loss during transcription and translation, constituting the DNA repair process.23
The mSASSS, BASFI, ASDAS, and Schober tests were scores to determine the disease activity of SpA in patients.24 Despite HLA-B*2704 and -B*2705 being the most commonly reported HLA alleles associated with SpA, the patients of Dr. Soetomo General Hospital with HLA-B*2704 and -B*2705 tended to show less severe disease when compared to patients with HLA-C and HLA-K. Cortes et al.5 found no correlation between radiographic severity and HLA-B*27, this is supported by the study from Arévalo et al.25 that revealed the HLA-B*27 negative patients had higher BASFI scores (higher severity). The study by Ruiz et al.26 also revealed that there was no association between the Schober score, which indicates low lumbar spine mobility, and the presence of HLA-B*27. However, these studies did not examine whether other HLA genes or subtypes were associated with SpA in HLA-B*27 negative patients. A study in Indonesia by Nasution et al. also concluded that HLA-B*27 lacked correlation with SpA in native Indonesia.14,25–27
Patients with HLA-C and HLA-K also have a short duration of illness, which indicates that with a shorter period, disease activity of SpA in HLA-C and HLA-K is excessively progressive. The HLA-C patients with a high prevalence of psoriasis in this study are aligned with the association between HLA-C and psoriasis that many studies have revealed.28–30 In line with a review by Akkoç et al., the ankylosing spondylitis (AS) patients with negative HLA-B*27 showed less correlation with psoriasis and inflammatory bowel disease.31 Although there are no studies on the association of HLA-C with the severity of SpA, the correlation of HLA-C with SpA-related diseases, especially psoriasis, has been widely discussed. On the other hand, the relation of HLA-K with SpA has never been reported in any studies.
The HLA-K is a pseudogene that commonly arises from deleterious mutations. Pseudogenes are usually not transcribed, but some pseudogenes can be transcribed but not translated or may also be translated into non-functional proteins.32 Frameshifts mutation and premature stop codons that trigger the emergence of pseudogenes cause the synthesis of entirely new or unusable proteins.33,34 Dysfunctional protein is suspected of disrupting the immune system, which makes the disease more severe. Non-functional protein from mutation can be misfolded at the endoplasmic reticulum (ER), thereby activating the unfolded protein response (UPR). The UPR that occurs in bone marrow macrophages leads to ER stress and causes increased production of IL-23, IFN-β, and IL-1α.35 A premature stop codon will cause a shortened protein that will be presented as an antigen resulting in an inflammation response.35,36 This may explain why HLA-K causes severe illness in patients.
Whereas the relationship between HLA types in this study was not statistically significant due to the small number of samples in each population, these results provide early evidence that further investigation is needed on the role of HLA types, especially HLA-K through SpA disease in Indonesia. A larger population is also needed for future research to clearly describe the association of the HLA type in Indonesian SpA patients.
The most common HLA types found in SpA patients at Dr. Soetomo were HLA-C (21.43%) and HLA-K (52.83%), with the most progressive disease activity indicated by poor mSASSS, BASFI, ASDAS, and Schober scores with a short duration of illness. While the type of HLA that is often associated with SpA, HLA-B*2704 and -B*2705, is only found in 12.86% and 1.43% of patients with less severe conditions. Larger-scale research is required to better understand the HLA type in SpA patients in Indonesia, which could be helpful for reliable SpA diagnosis and treatment in the future.
Figshare: Underlying data for ‘HLA class I and discoveries of the HLA-K (pseudogene) related to disease severity and progression in patients with spondyloarthritis in Dr. Soetomo General Hospital, a tertiary health care center in Surabaya, Indonesia’, https://doi.org/10.6084/m9.figshare.20250585.
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0)
NCBI Protein: Homo sapiens isolate YUL1 MHC class I antigen (HLA-B). Accession number ON986008; https://identifiers.org/ncbiprotein:ON986008
NCBI Protein: Homo sapiens isolate YUL2 MHC class I antigen (HLA-B). Accession number ON986009; https://identifiers.org/ncbiprotein:ON986009
NCBI Protein: Homo sapiens isolate YUL9 MHC class I antigen (HLA-B) HLA-B*18 allele. Accession number ON986010; https://identifiers.org/ncbiprotein:ON986010
NCBI Protein: Homo sapiens isolate YUL12 MHC class I antigen (HLA-B) HLA-B*27:06 allele. Accession number ON986011; https://identifiers.org/ncbiprotein:ON986011
NCBI Protein: Homo sapiens isolate YUL13 MHC class I antigen (HLA-B). Accession number ON986012; https://identifiers.org/ncbiprotein:ON986012
NCBI Protein: Homo sapiens isolate YUL15 MHC class I antigen (HLA-B) HLA-B*35 allele. Accession number ON986013; https://identifiers.org/ncbiprotein:ON986013
NCBI Protein: Homo sapiens isolate YUL27 MHC class I antigen (HLA-B). Accession number ON986014; https://identifiers.org/ncbiprotein:ON986014
NCBI Protein: Homo sapiens isolate YUL32 MHC class I antigen (HLA-B) HLA-B*18 allele. Accession number ON986015; https://identifiers.org/ncbiprotein:ON986015
NCBI Protein: Homo sapiens isolate YUL33 MHC class I antigen (HLA-B). Accession number ON986016; https://identifiers.org/ncbiprotein:ON986016
NCBI Protein: Homo sapiens isolate YUL36 MHC class I antigen (HLA-B). Accession number ON986017; https://identifiers.org/ncbiprotein:ON986017
NCBI Protein: Homo sapiens isolate YUL37 MHC class I antigen (HLA-B). Accession number ON986018; https://identifiers.org/ncbiprotein:ON986018
NCBI Protein: Homo sapiens isolate YUL41 MHC class I antigen (HLA-B). Accession number ON986019; https://identifiers.org/ncbiprotein:ON986019
NCBI Protein: Homo sapiens isolate YUL47 MHC class I antigen (HLA-B) HLA-B*18 allele. Accession number ON986020; https://identifiers.org/ncbiprotein:ON986020
NCBI Protein: Homo sapiens isolate YUL54 MHC class I antigen (HLA-B) HLA-B*35 allele. Accession number ON986021; https://identifiers.org/ncbiprotein:ON986021
NCBI Protein: Homo sapiens isolate YUL59 MHC class I antigen (HLA-B) HLA-B*27:05 allele. Accession number ON986022; https://identifiers.org/ncbiprotein:ON986022
NCBI Protein: Homo sapiens isolate YUL62 MHC class I antigen (HLA-B). Accession number ON986023; https://identifiers.org/ncbiprotein:ON986023
NCBI Protein: Homo sapiens isolate YUL63 MHC class I antigen (HLA-B) HLA-B*57 allele. Accession number ON986024; https://identifiers.org/ncbiprotein:ON986024
NCBI Protein: Homo sapiens YUL4 MHC class I antigen (HLA-K) pseudogene. Accession number ON986025; https://identifiers.org/ncbiprotein:ON986025
NCBI Protein: Homo sapiens YUL5 MHC class I antigen (HLA-K) pseudogene. Accession number ON986026; https://identifiers.org/ncbiprotein:ON986026
NCBI Protein: Homo sapiens YUL6 MHC class I antigen (HLA-K) pseudogene. Accession number ON986027; https://identifiers.org/ncbiprotein:ON986027
NCBI Protein: Homo sapiens YUL8 MHC class I antigen (HLA-K) pseudogene. Accession number ON986028; https://identifiers.org/ncbiprotein:ON986028
NCBI Protein: Homo sapiens YUL14 MHC class I antigen (HLA-K) pseudogene. Accession number ON986029; https://identifiers.org/ncbiprotein:ON986029
NCBI Protein: Homo sapiens YUL16 MHC class I antigen (HLA-K) pseudogene. Accession number ON986030; https://identifiers.org/ncbiprotein:ON986030
NCBI Protein: Homo sapiens YUL17 MHC class I antigen (HLA-K) pseudogene. Accession number ON986031; https://identifiers.org/ncbiprotein:ON986031
NCBI Protein: Homo sapiens YUL18 MHC class I antigen (HLA-K) pseudogene. Accession number ON986032; https://identifiers.org/ncbiprotein:ON986032
NCBI Protein: Homo sapiens YUL20 MHC class I antigen (HLA-K) pseudogene. Accession number ON986033; https://identifiers.org/ncbiprotein:ON986033
NCBI Protein: Homo sapiens YUL21 MHC class I antigen (HLA-K) pseudogene. Accession number ON986034; https://identifiers.org/ncbiprotein:ON986034
NCBI Protein: Homo sapiens YUL22 MHC class I antigen (HLA-K) pseudogene. Accession number ON986035; https://identifiers.org/ncbiprotein:ON986035
NCBI Protein: Homo sapiens YUL24 MHC class I antigen (HLA-K) pseudogene. Accession number ON986036; https://identifiers.org/ncbiprotein:ON986036
NCBI Protein: Homo sapiens YUL26 MHC class I antigen (HLA-K) pseudogene. Accession number ON986037; https://identifiers.org/ncbiprotein:ON986037
NCBI Protein: Homo sapiens YUL26 MHC class I antigen (HLA-K) pseudogene. Accession number ON986038; https://identifiers.org/ncbiprotein:ON986038
NCBI Protein: Homo sapiens YUL29 MHC class I antigen (HLA-K) pseudogene. Accession number ON986039; https://identifiers.org/ncbiprotein:ON986039
NCBI Protein: Homo sapiens YUL31 MHC class I antigen (HLA-K) pseudogene. Accession number ON986040; https://identifiers.org/ncbiprotein:ON986040
NCBI Protein: Homo sapiens YUL34 MHC class I antigen (HLA-K) pseudogene. Accession number ON986041; https://identifiers.org/ncbiprotein:ON986041
NCBI Protein: Homo sapiens YUL35 MHC class I antigen (HLA-K) pseudogene. Accession number ON986042; https://identifiers.org/ncbiprotein:ON986042
NCBI Protein: Homo sapiens YUL39 MHC class I antigen (HLA-K) pseudogene. Accession number ON986043; https://identifiers.org/ncbiprotein:ON986043
NCBI Protein: Homo sapiens YUL42 MHC class I antigen (HLA-K) pseudogene. Accession number ON986044; https://identifiers.org/ncbiprotein:ON986044
NCBI Protein: Homo sapiens YUL43 MHC class I antigen (HLA-K) pseudogene. Accession number ON986045; https://identifiers.org/ncbiprotein:ON986045
NCBI Protein: Homo sapiens YUL45 MHC class I antigen (HLA-K) pseudogene. Accession number ON986046; https://identifiers.org/ncbiprotein:ON986046
NCBI Protein: Homo sapiens YUL46 MHC class I antigen (HLA-K) pseudogene. Accession number ON986047; https://identifiers.org/ncbiprotein:ON986047
NCBI Protein: Homo sapiens YUL48 MHC class I antigen (HLA-K) pseudogene. Accession number ON986048; https://identifiers.org/ncbiprotein:ON986048
NCBI Protein: Homo sapiens YUL49 MHC class I antigen (HLA-K) pseudogene. Accession number ON986049; https://identifiers.org/ncbiprotein:ON986049
NCBI Protein: Homo sapiens YUL50 MHC class I antigen (HLA-K) pseudogene. Accession number ON986050; https://identifiers.org/ncbiprotein:ON986050
NCBI Protein: Homo sapiens YUL52 MHC class I antigen (HLA-K) pseudogene. Accession number ON986051; https://identifiers.org/ncbiprotein:ON986051
NCBI Protein: Homo sapiens YUL53 MHC class I antigen (HLA-K) pseudogene. Accession number ON986052; https://identifiers.org/ncbiprotein:ON986052
NCBI Protein: Homo sapiens YUL56 MHC class I antigen (HLA-K) pseudogene. Accession number ON986053; https://identifiers.org/ncbiprotein:ON986053
NCBI Protein: Homo sapiens YUL60 MHC class I antigen (HLA-K) pseudogene. Accession number ON986054; https://identifiers.org/ncbiprotein:ON986054
NCBI Protein: Homo sapiens YUL61 MHC class I antigen (HLA-K) pseudogene. Accession number ON986055; https://identifiers.org/ncbiprotein:ON986055
NCBI Protein: Homo sapiens YUL64 MHC class I antigen (HLA-K) pseudogene. Accession number ON986056; https://identifiers.org/ncbiprotein:ON986056
NCBI Protein: Homo sapiens YUL65 MHC class I antigen (HLA-K) pseudogene. Accession number ON986057; https://identifiers.org/ncbiprotein:ON986057
NCBI Protein: Homo sapiens YUL66 MHC class I antigen (HLA-K) pseudogene. Accession number ON986058; https://identifiers.org/ncbiprotein:ON986058
NCBI Protein: Homo sapiens YUL68 MHC class I antigen (HLA-K) pseudogene. Accession number ON986059; https://identifiers.org/ncbiprotein:ON986059
NCBI Protein: Homo sapiens YUL69 MHC class I antigen (HLA-K) pseudogene. Accession number ON986060; https://identifiers.org/ncbiprotein:ON986060
NCBI Protein: Homo sapiens YUL70 MHC class I antigen (HLA-K) pseudogene. Accession number ON986061; https://identifiers.org/ncbiprotein:ON986061
NCBI Protein: Homo sapiens isolate YUL3 MHC class I antigen (HLA-C) HLA-C*06 allele. Accession number ON986062; https://identifiers.org/ncbiprotein:ON986062
NCBI Protein: Homo sapiens isolate YUL7 MHC class I antigen (HLA-C) HLA-C*08 allele. Accession number ON986063; https://identifiers.org/ncbiprotein:ON986063
NCBI Protein: Homo sapiens isolate YUL10 MHC class I antigen (HLA-C) HLA-C*16 allele. Accession number ON986064; https://identifiers.org/ncbiprotein:ON986064
NCBI Protein: Homo sapiens isolate YUL11 MHC class I antigen (HLA-C) HLA-C*04 allele. Accession number ON986065; https://identifiers.org/ncbiprotein:ON986065
NCBI Protein: Homo sapiens isolate YUL19 MHC class I antigen (HLA-C) HLA-C*16 allele. Accession number ON986066; https://identifiers.org/ncbiprotein:ON986066
NCBI Protein: Homo sapiens isolate YUL23 MHC class I antigen (HLA-C) HLA-C*04 allele. Accession number ON986067; https://identifiers.org/ncbiprotein:ON986067
NCBI Protein: Homo sapiens isolate YUL25 MHC class I antigen (HLA-C) HLA-C*08 allele. Accession number ON986068; https://identifiers.org/ncbiprotein:ON986068
NCBI Protein: Homo sapiens isolate YUL30 MHC class I antigen (HLA-C) HLA-C*08 allele. Accession number ON986069; https://identifiers.org/ncbiprotein:ON986069
NCBI Protein: Homo sapiens isolate YUL38 MHC class I antigen (HLA-C) HLA-C*16 allele. Accession number ON986070; https://identifiers.org/ncbiprotein:ON986070
NCBI Protein: Homo sapiens isolate YUL40 MHC class I antigen (HLA-C) HLA-C*08 allele. Accession number ON986071; https://identifiers.org/ncbiprotein:ON986071
NCBI Protein: Homo sapiens isolate YUL44 MHC class I antigen (HLA-C) HLA-C*08 allele. Accession number ON986072; https://identifiers.org/ncbiprotein:ON986072
NCBI Protein: Homo sapiens isolate YUL51 MHC class I antigen (HLA-C) HLA-C*08 allele. Accession number ON986073; https://identifiers.org/ncbiprotein:ON986073
NCBI Protein: Homo sapiens isolate YUL55 MHC class I antigen (HLA-C) HLA-C*08 allele. Accession number ON986074; https://identifiers.org/ncbiprotein:ON986074
NCBI Protein: Homo sapiens isolate YUL57 MHC class I antigen (HLA-C) HLA-C*06 allele. Accession number ON986075; https://identifiers.org/ncbiprotein:ON986075
NCBI Protein: Homo sapiens isolate YUL58 MHC class I antigen (HLA-C) HLA-C*08 allele. Accession number ON986076; https://identifiers.org/ncbiprotein:ON986076
NCBI Protein: Homo sapiens isolate YUL67 MHC class I antigen (HLA-C) HLA-C*08 allele. Accession number ON986077; https://identifiers.org/ncbiprotein:ON986077
Written informed consent for publication of the patients’ details was obtained from the patients.
The authors acknowledge the study participants and the hospital staff for their time, dedication, and contribution to this study.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
No
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
No
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: HLA immunogenetics
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Spondyloarthritis
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 07 Sep 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)