Keywords
European Nucleotide Archive, submission tools, validation functions, specimen voucher, culture collection, bio material, NCBI Biocollections
This article is included in the Bioinformatics gateway.
This article is included in the EMBL-EBI collection.
European Nucleotide Archive, submission tools, validation functions, specimen voucher, culture collection, bio material, NCBI Biocollections
The generation and archiving of sequence data and associated metadata at large scale have transformed and promoted research in the life sciences. Sequence data have been essential to scientific breakthroughs in several fields such as medicine, food security, evolutionary biology and biodiversity conservation.
One of the most important aspects of the archiving of sequence data is metadata management, which is crucial for the accurate description of Earth’s genetic and genomic biodiversity and its preservation in molecular sequence collections (Waterhouse et al. 2021). By holding enriched metadata, such as biological source attributes, that describe the material provenance of sequence data (allowing linking to the specimen of origin), molecular sequence collections facilitate connections between molecular biology, taxonomy, systematics and biodiversity research, increasing the discoverability and usability of data by researchers worldwide.
The infrastructure for storing and sharing of sequence data is the International Nucleotide Sequence Database Collaboration (INSDC, Arita et al. 2021) that operates between the DNA Data Bank of Japan (DDBJ, Fukuda et al. 2021), the National Centre for Biotechnology Information (NCBI, Sayers et al. 2021) and the European Nucleotide Archive (ENA, Cummins et al. 2022) that stands as its European node. The INSDC contains currently over 236 million sequences and 1.7 billion reads (https://www.ncbi.nlm.nih.gov/genbank/statistics/), holding a large body of associated metadata related to sequenced sample sources, as culture collection or natural history collection annotations. However, for a number of records these metadata may be incomplete or ambiguous, which hinders the linking of the sequence data to their origin and therefore reduces data reusability.
The Biodiversity Community Integrated Knowledge Library (BiCIKL) is a Horizon 2020 project that aims to establish open science practices in the biodiversity domain by providing Findable, Accessible, Interoperable and Reusable (FAIR) access and developing new methods and workflows for linking data along the biodiversity research cycle, namely from data resources of molecular biology, natural history collections, taxonomy, and literature (Penev et al. 2022). To take full advantage of these workflows a foundation of well-structured and accessible metadata is required, namely in the molecular sequence databases. Therefore, in the scope of this project we have developed a tool for driving the accurate and complete reporting of biological source metadata: the ENA Source Attribute Helper.
The metadata that refers to the biological source of sequence data is described for sequences in the source feature qualifiers that are embedded in the sequence flat files (INSDC 2021, https://www.ebi.ac.uk/ena/WebFeat/) and in the samples’ attributes. These will be hereafter referred simply as ‘attributes’. These attributes are submitted to the ENA at the time of data deposition or in subsequent updates. ENA holds many routes for data submission (such as web interfaces, RESTful Application Programming Interfaces (APIs) and locally installed command-line tools), so there is no single point of entry for supporting submission of accurate provenance metadata. Therefore, the Source Attribute Helper is a publicly accessible open-source tool that may be used as a free-standing service independently across platforms and workflows. The initial version of the tool focuses on the sequence and sample attributes that identify the specimen, culture, or material from which the sequence was derived, namely /specimen_voucher, /culture_collection, and /bio_material. These attributes are formatted according to the Darwin Core Standards (Wieczorek et al. 2012) and follow a Darwin Core Triplet format, composed of Institution code, collection code and the specimen, culture, or material id, accordingly (Table 1). The tool was developed to help users fetch accurate information regarding the institution and collections codes of the specimen, culture, or material, and construct and validate the string to be submitted as an attribute of the sequence data. The tool does not however support the search of voucher specimen codes, as these need to be obtained directly from the institutions.
The value of the attributes follows a Darwin Core triplet format (according to Darwin Core Standards, Wieczorek et al. 2012). Specimen ID, culture ID, and material ID are mandatory values. Institution code is optional for specimen voucher and bio material, but mandatory for culture collection. Collection code is always optional. When collection code is provided, institution code is mandatory (see INSDC 2021, also available at https://www.ebi.ac.uk/ena/WebFeat/, for more details on these attributes). ENA, European Nucleotide Archive; API, Application Programming Interface; INSDC, International Nucleotide Sequence Database Collaboration.
In this paper we describe the design and implementation of the API. We also describe use cases for its application, highlighting its utility for increasing the accuracy of biological source attributes in molecular databases and promoting reusability.
The ENA Source Attribute Helper, although described as a single tool, comprises several endpoints with different functions, namely, to display metadata associated with institutions or collections, to validate the attribute string provided by the user according to the institutions and collections database, and to construct the attribute string based on data input by the user. The code is available from GitHub and is archived with Zenodo (Jayathilaka & Gupta 2022).
For the retrieval of information on the institutions and collections and subsequent validation, the application uses the data available in the NCBI Biocollections (RRID:SCR_016459). NCBI Biocollections is a curated database of metadata for herbaria, museums, culture collections, and other natural history collections, that are connected to records in INSDC, and is maintained by the NCBI taxonomy group (Sharma et al. 2018). It includes institution and collection codes and their URLs, where available, that allow users to find additional information. New records are added to the database upon submission of information together with sequence records to INSDC. The ENA Source Attribute Helper consumes the curated data from NCBI Biocollections to get, validate and construct the values for the attributes (Tables 2 and 3). Currently, the available Biocollections database files are retrieved manually from the ftp server and imported into the ENA ElasticSearch datastore.
NCBI, National Centre for Biotechnology Information; ENA, European Nucleotide Archive.
| Field | Example | Type | Requirement | Notes |
|---|---|---|---|---|
| _id | x-P-TIEBpiSBteIpqVU0 | System set uuid | System set | System set uuid. |
| inst_id | 1111 | Integer | Mandatory | Provided Institution id |
| inst_code | CAMZM | String | Mandatory | Institution Code |
| unique_name | UMZC | String | Mandatory | Institution Unique Name |
| synonyms | AEIC | String | Optional | Institute Synonym |
| inst_name | University Museum of Zoology Cambridge | String | Mandatory | Name of the Institution |
| country | United Kingdom | String | Mandatory | Institution Country |
| address | Downing Street, Cambridge, CB2 3EJ, Cambridge | String | Mandatory | Address of the Institution |
| collection_type | museum | String | Mandatory | Type of Collection |
| qualifier_type | specimen_voucher | String | Mandatory | Attribute: specimen_voucher/bio_material/culture_collection |
| home_url | http://www.zoo.cam.ac.uk/museum/ | String | Optional | Home Page URL |
| url_rule | String | Optional | URL Rule Page |
NCBI, National Centre for Biotechnology Information; ENA, European Nucleotide Archive.
| Field | Example | Type | Requirement | Notes |
|---|---|---|---|---|
| _id | dee11d4e-63c6-4d90-983c-5c9f1e79e96c | System set uuid | System set | System set uuid |
| coll_id | 222 | Integer | Mandatory | Provided Collection id |
| inst_id | 12345 | Integer | Mandatory | Mapped Institution id |
| coll_code | Annelid | String | Mandatory | Collection Code |
| coll_name | Annelid Collection | String | Mandatory | Name of the Collection |
| coll_type | museum | String | Mandatory | Type of Collection |
| qualifier_type | specimen_voucher | String | Mandatory | Attribute: specimen_voucher/bio_material/culture_collection |
| coll_url | http://nature.ca/collections/inverts_e.cfm | String | Optional | Collection Reference URL |
| coll_url_rule | https://science.mnhn.fr/institution/mnhn/collection/ar/item/ar | String | Optional | Collection URL Rule |
The development of the ENA Source Attribute Helper API was based on the following tools and frameworks:
1. Spring Boot API framework: This is a framework used for building RESTFul APIs that are accessible from various platforms/clients including but not limited to web browsers, mobile devices, desktop applications etc.
2. ElasticSearch datastore: The application utilises strengths of ElasticSearch datastore to enable text search over JavaScript Object Notation (JSON) data over multiple properties and provide suggestions/similar matches.
3. Spring Data and object–relational mapping (ORM): The application utilises Spring Data libraries to create abstractions over repository and custom object mappings along with usage of an ORM (like Hibernate).
4. Postman: This is an open-source tool for testing, monitoring and publishing APIs.
5. Swagger: This is an Interface Description Language (IDL) used for the description of RESTful APIs. It allows the visualisation of the various API endpoints and an easy execution of the commands.
Construct & validation flows
The main function of the API is to validate and construct the Attribute Values (see Table 1), to ensure that these are aligned with the format definition. The user inputs a code or name (at least one character) for the institution or collection and the application suggests the closest options available, so that the user can select the correct option. The construct and validation flows are represented in Figure 1.
Core
The core of the application is built using the Spring Boot framework, which follows a layered architecture approach (Presentation Layer, Business Layer, Persistence Layer, Database Layer) in which each layer communicates to other layers in a hierarchical order. The Database entities (ElasticSearch datastore) and the Backend APIs (Web API Layer) are represented in Figure 2.
API endpoints
API endpoints are the channels through which other applications can communicate with or consume an API. They are represented by Universal Resource Locators (URL), which serve as points of entry.
Table 4 describes the various endpoints that this API provides. The endpoint to get error-codes is an additional endpoint that allows users to fetch the definitions of the error codes, which may be returned by the system. This may be useful for system integration and error handling on the client side.
ENA, European Nucleotide Archive; API, Application Programming Interface.
API access & tools
The API can be accessed using:
• any Web Browser
• any scripting/programming language-based REST client
• command line tools like cURL and Wget
• testing tools like Swagger user interface (UI) and Postman
Testing tools, such as the Swagger UI, facilitate the usage of the API even by a non-technical person (Figure 3), while other tools such as Postman may require a higher level of technical understanding to know how to consume the API.
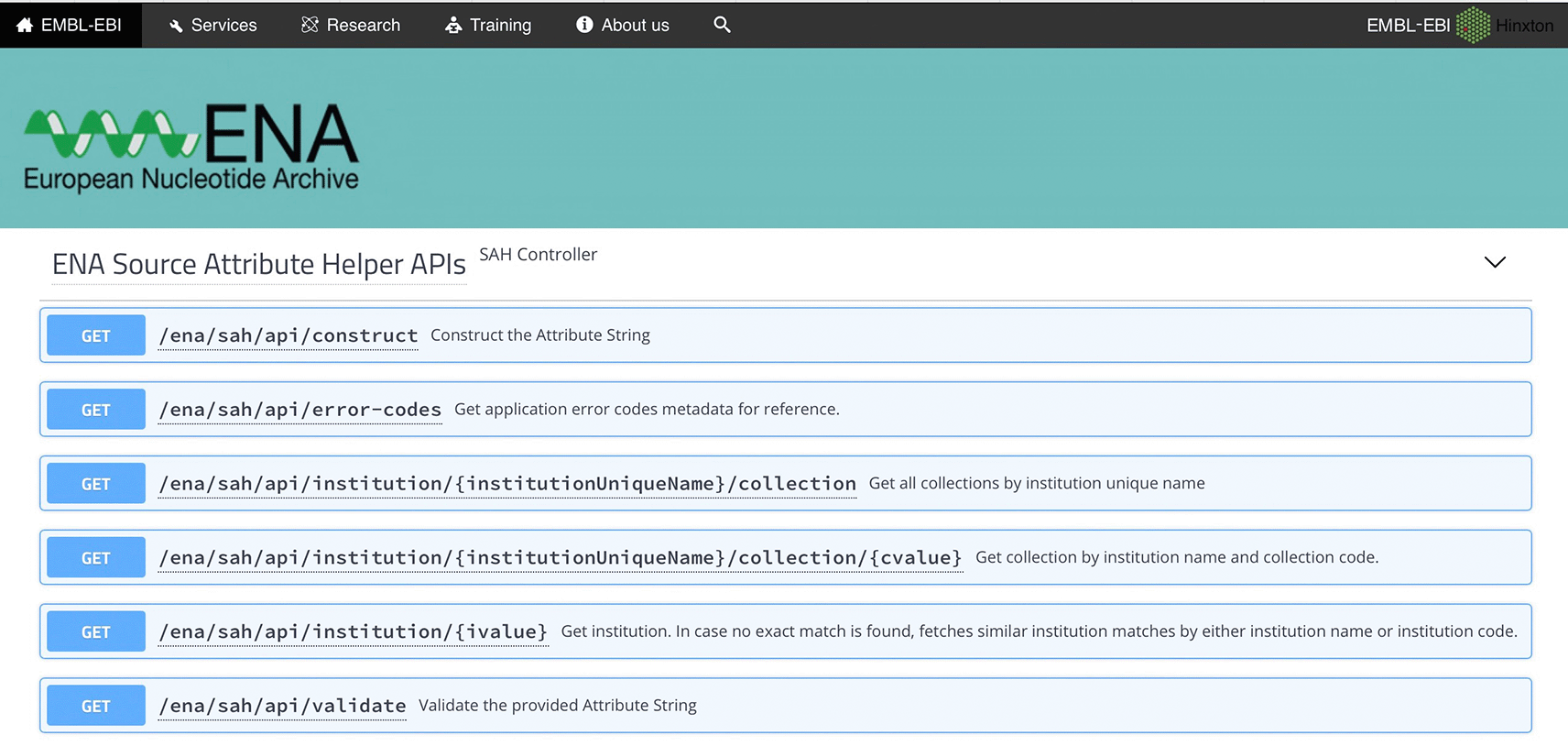
UI, User Interface; ENA, European Nucleotide Archive; API, Application Programming Interface.
Using the Swagger interface
Swagger is a web browser based graphical UI that provides a set of form fields and hints for interacting with a RESTful API. It helps a user to interact with and test the API by hiding the complexity of building correct requests. It uses annotations and descriptions in the source code of the application to describe the API in human readable format. Figure 3 shows the general UI for Swagger, and an example of a JSON response for the /validate endpoint (which validates the given attribute string) is displayed in Figure 4. Figure 5 shows an example of a JSON response for the /construct endpoint, which validates input and constructs the attribute string based on the provided parameters.
Using the cURL command-line tool
cURL is a widely available free and open-source command-line tool for transferring data using URL syntax. Figures 6 and 7 show two examples of the usage of the /validate and /construct API endpoints to validate and construct the attribute string, respectively, using cURL.
Using the Postman API client
The Postman API client is a tool to easily explore, debug, and test APIs while also enabling users to define complex API requests for HTTP, REST, SOAP, GraphQL, and WebSockets. In the development of the tool, we engaged it to inspect API endpoints and their responses. An example JSON response for the /validate endpoint, which validates a given attribute string and presents the support data for the institution and collection values, is shown in Figure 8.
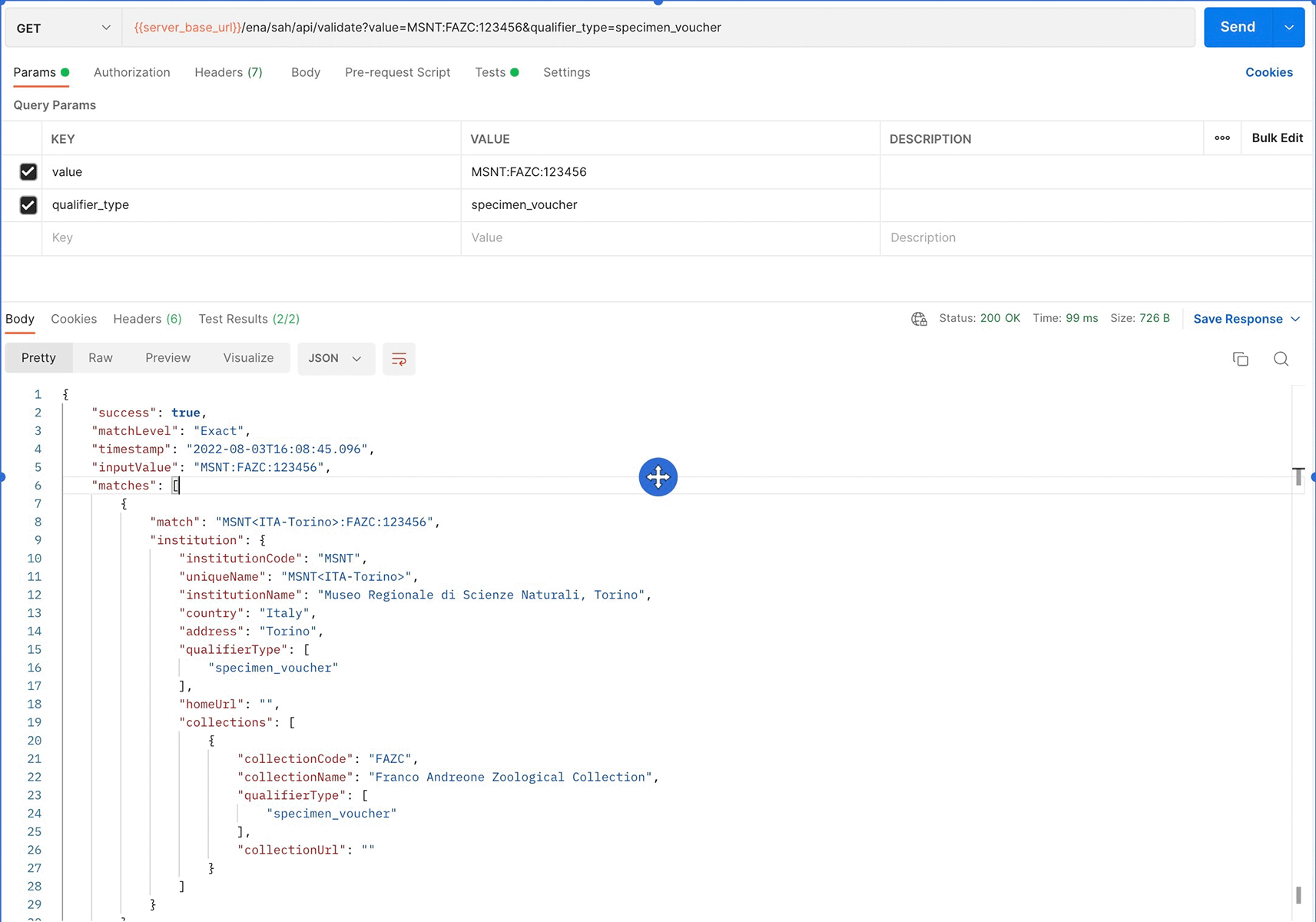
UI, User Interface.
Other API clients - Python
Python is a scripting/programming language that allows a quick output and integrates systems more effectively. Figures 9 and 10 show basic code examples to demonstrate querying two of the available API endpoints - /validate and /construct.
Users submitting sequence related data may need to look for the unique code for the institution holding the voucher associated with the data. The API endpoint Get Institution allows the user to fetch the Institution details by providing either the institution name or code (Table 5). The type of attribute (‘qualifier_type’: specimen voucher, culture collection, or bio material) may also be optionally specified, but if none is provided the API will search within all attributes. The API searches both for exact matches and for partial matches. The API response will include the metadata for all institutions with exact or partial matches to the input value, allowing the user to confirm the details of the institution that is holding the voucher.
Requirements and an example are also provided. API, Application Programming Interface.
Once the user knows the unique institution code for their voucher, they may need to identify the unique code for the collection to input in the attribute string. The API has two endpoints that allow users to search for the collection codes. In both endpoints the type of attribute (‘qualifier_type’: specimen voucher, culture collection, or bio material) may be optionally specified.
The endpoint Get Collections for the Institution allows to fetch all collections in a given institution by providing the institution’s unique name (unique code, Table 6). This operation looks only for an exact match of the institution's unique name and returns the complete list of collections within that institution and associated metadata. If the institution’s unique name is not found in the database the endpoint does not return any record.
Requirements and an example are also provided. API, Application Programming Interface.
The endpoint Get Collections by Institution Unique name and Collection Code allows users to obtain the metadata of a given collection of an institution by providing the institution’s unique name (unique code) and known collection code (Table 7). This endpoint searches for an exact match of the institution's unique name, and a full or partial match of the collection code and returns the metadata for the collection found. If the institution’s unique name or collection code are not found in the database, the endpoint does not return any record.
Requirements and an example are also provided. API, Application Programming Interface.
Users that are already aware of the format of the biological source attributes, and have information about the institution and collection codes, may use the API endpoint Validate Attribute to validate the attribute string. The user needs to provide the attribute string in the format detailed in Table 1, according to the attribute type. The attribute type (‘qualifier_type’: specimen voucher, culture collection, or bio material) may also be specified to narrow the search, but if none is provided the API will search within all attribute values (Table 8). The API performs the search for the exact match, but if none is found, a search for partial matches for the provided string will be performed. The response includes the type of match (match level exact or partial), a recommendation for the qualifier value (match) that may correspond to the input or include corrections to the unique values of the institution and collections, and the metadata of the referred institution and collections (Figure 8). If the match isn’t exact and there is more than one possible match to the attribute string input by the user, the response will include all possible matches and associated metadata.
Requirements and an example are also provided. API, Application Programming Interface.
| Parameters | Example | Type | Requirement | Notes |
|---|---|---|---|---|
| value | MSNT:FAZC:123456 | String | Mandatory | Attribute string to be validated in the format described in Table 1 |
| qualifier_type | specimen_voucher, bio_material, culture_collection | String | Optional | Filters results for a specific attribute |
Users may use the API endpoint Construct the Attribute to help them obtain the correct attribute string for referring to the biological source of the voucher linked with the sequence data. In this endpoint the user needs to provide separately the expected values for the institution, collection, and the ID of the specimen, culture, or material, depending on the attribute type (Table 9). The type of attribute (‘qualifier_type’: specimen voucher, culture collection, or bio material) may also be specified to narrow the search, but if none is provided the API will search within all attribute values. As in the validate function, the API also searches for partial matches. The response includes the type of match (match level exact or partial), the constructed attribute string from the values input by the user (input value), a recommendation for the attribute string (match) that may correspond to the input or include corrections to the unique values of the institution and collections, and the metadata of the referred institution and collections. In Figure 7 (cURL request for /construct API endpoint) we can see a case where the match is only partial, as there is more than one option for the institution code provided by the user. In these situations, the response will include all possible matches and associated metadata.
Requirements and an example are also provided. API, Application Programming Interface.
Further developments of the ENA Source Attribute Helper API are planned.
Regarding the retrieval of the data, an automated flow for getting the updated files from the NCBI servers regularly is planned for implementation.
The development of a Graphical User Interface (GUI) is also planned for implementation, likely embedded in, or accessible from, one or more of ENA's existing submission tools. This will allow more intuitive searches for Institutions and/or Collections metadata and the validation/construction of the qualifier values to be more accessible to inexperienced users. This UI will connect to the API to support features like:
Considering the increasing rates of generation and submission of sequence data to public repositories it becomes increasingly important to assure the greatest accuracy and precision of associated metadata. Hence, we have developed and deployed a tool that will considerably help users to provide accurate metadata for reference to the biological source of sequence data. We have described the ENA Source Attribute Helper API design and implementation and discussed its main usages. We expect this tool to promote and support the submission of better structured and more richly described data that will provide a stronger foundation to strengthen the value of natural history collections, taxonomic expertise, and biodiversity knowledge.
For biodiversity research, the wider availability of correctly structured biological source attributes in sequence data will, for instance, improve the linkage with distribution data in the Global Biodiversity Information Facility (GBIF; GBIF 2022). GBIF holds a data-clustering feature that identifies records that are potentially related by matching similar metadata entries (GBIF 2020). In the case of the INSDC Sequences dataset in GBIF the fields used for matching are the biological source attributes in the triple Darwin Core format (Grosjean & Robertson 2021). Therefore, we expect that the number of sequence records linked to specimens in natural history collections and to their distribution data will increase with the usage of the ENA Source Attribute Helper API. Monitoring these links will help us to measure the impact of the usage of this tool.
Overall, we expect the enrichment of the provenance metadata of sequences in molecular biology repositories to contribute to boost our understanding of, and effectiveness of response to global challenges such as biodiversity loss, ecosystem change and food security.
The data used in this API are available at https://www.ncbi.nlm.nih.gov/biocollections. The API retrieves data from the institutions, collections and unique institutions codes files that are available for public access at https://ftp.ncbi.nih.gov/pub/taxonomy/biocollections/.
Software available from: https://www.ebi.ac.uk/ena/sah/api/
Source code available from: https://github.com/enasequence/ena-source-annotation-helper
Archived source code at time of publication: https://doi.org/10.5281/zenodo.7063227 (Jayathilaka & Gupta 2022)
License: Apache License 2.0 license
We would like to thank Conrad Schoch, Shobba Sharma and the NCBI taxonomy team for providing information on the NCBI biocollections database and its ftp access. We would also like to acknowledge Mathias Dillen, Marcus Ernst, Quentin Groom, Anton Güntsch and Tim Robertson for their inputs to an earlier version of the manuscript and to the BiCIKL Consortium for their support.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genomics and NGS
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bioinformatics, Software Design, Biodiversity Informatics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 13 Sep 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)