Keywords
Agronomic traits, machine learning, multi-omics, plant improvement
This article is included in the Plant Science gateway.
This article is included in the Artificial Intelligence and Machine Learning gateway.
This article is included in the Plant Computational and Quantitative Genomics collection.
Agronomic traits, machine learning, multi-omics, plant improvement
The global agricultural system is threatened by ecological events such as climate change and other environmental stresses.1,2 These events affect the yield, stability, and quality of production of economically important plants, i.e., medicinal plants, fruit crops, food crops, cereal or grain crops, legume seed crops or pulses, etc.1,3 These challenges are addressed by omics approaches via numerous unconventional improvement methodologies.4,5 Omics approaches involve analysis of the constituents of genome sequence and other macromolecules generated from encoded genomic information. The utilization of omics-derived knowledge and technologies in plant improvement strategies is limited and difficult. Other drawbacks of omics technologies include a lack of data integration and effective phenotype-genotype correlation strategies. As a result, promoting the integration of computational biology and plant genomics to assist plant development is critical.6,7 This paper gives an overview of omics methodologies for improving plant agronomic traits as well as essential curated plant genomic data sources. We discuss the bioinformatics software, tools, and packages that are utilized in omics-based plant improvement research. We also dissect how machine learning algorithms has been used to improve agronomic features of commercially significant plants, their major contributions and future outlook in plant omics and agronomics.
Genome sequencing was made possible with the advent of sequencing technologies.8,9 Complete sequencing of a plant genome was first demonstrated for a model plant named Arabidopsis (Arabidopsis thaliana)10,11 and afterward for rice (Oryza sativa). Subsequently, the whole genome of over 250 species in the plant kingdom have been sequenced: bryophytes, pteridophytes, gymnosperms, and angiosperms12,13 (Figure 1). Angiosperms account for 95% of the sequenced species, most of which are economically important plants or their wild relatives (Figure 2). Food crops like rice, wheat, beans, oat, maize, and soybean are among the sequenced plants, as are ornamental plants like orchid and hibiscus, industrial plants like oilseed, hemp, and spice/herbs like garlic, ginger, turmeric, moringa, artemisia, and neem, which are known for their high therapeutic value.

Most sequenced plants are angiosperms and are subdivided into three groups. Most of the sequenced angiosperms fall under rosids and asterids clades. Other sequenced angiosperms clades are grouped here as other dicots.

94% of sequenced plants are angiosperms consisting of both monocots and dicots. Percentage of other plants are 1%, 2%, 3% for Pteridophytes, Bryophytes, and Gymnosperms, respectively.
There have been a variety of databases created to access plant genome datasets.14,11 The model plant A. thaliana genome database launched in 2001 was the premier plant genome database.10,15 Subsequently, many databases and resources have been developed for plant genomes. The earliest genome databases were essentially archives of genome sequence data. These databases have expanded into genome portals/hubs that combine different genomic data and web servers that offer online genomics analysis. The availability of annotated plant genome data has led to many discoveries, including genome organization and gene function.16 These discoveries elucidate the complexity, evolution, and dynamics of plant genomes, contributing to a deeper understanding of plant biology.6,17 Available genomic information includes cis-elements, gene expression data, protein interactome, transcriptional and post-transcriptional data. These genome databases exist as single species and comprehensive databases as shown in Table 1.
| Database | Description | Website |
|---|---|---|
| AgBase | A unified resource for functional analysis in agriculture | http://www.agbase.msstate.edu/ |
| Ensembl Plants | A genome-centric portal for plant species | http://plants.ensembl.org |
| AutoSNPdb | An annotated single nucleotide polymorphism database for crop plants | http://autosnpdb.qfab.org.au/ |
| BarleyBase | An expression profiling database for plant genomics | http://www.barleybase.org/ |
| CR-EST | A resource for crop ESTs Search for sequence, classification, clustering and annotation data of crop EST projects | http://pgrc.ipk-gatersleben.de/cr-est/ |
| CSRDB | A small RNA integrated database and browser resource for cereals | http://sundarlab.ucdavis.edu/smrnas/ |
| ChromDB | The Chromatin Database | http://www.chromdb.org/ |
| DRASTICx97INSIGHTS | Querying information in a plant gene expression database | http://www.drastic.org.uk/ |
| FLAGdb++ | A Database for the Functional Analysis of the Arabidopsis Genome | http://urgv.evry.inra.fr/projects/FLAGdb++/HTML/index.shtml |
| GCP | The Generation Challenge Programme | http://www.generationcp.org/ |
| GGT | Graphical GenoTypes - Software for visualization and analysis of genetic data | http://www.plantbreeding.wur.nl/ |
| GabiPD | Integrative Plant Omics Database | http://www.gabipd.org/ |
| GeneSeqer@PlantGDB | Gene structure prediction in plant genomes - Predict gene structures of plant genomes | http://www.plantgdb.org/PlantGDB-cgi/GeneSeqer/PlantGDBgs.cgi |
| GrainGenes | The genome database for small-grain crops | http://wheat.pw.usda.gov/index.shtml |
| Gramene | A resource for comparative grass genomics | http://www.gramene.org/ |
| MIPS | Analysis and annotation of genome information | http://mips.gsf.de/ |
| MetaCrop | A detailed database of crop plant metabolism | http://metacrop.ipk-gatersleben.de/ |
| NIASGBdb | National Institute of Agrobiological Sciences Gene Bank DataBase | http://www.gene.affrc.go.jp/databases_en.php |
| P3DB | Plant Protein Phosphorylation Database | http://digbio.missouri.edu/p3db/ |
| PHYTOPROT | A Database of Clusters of Plant Proteins | https://urgi.versailles.inra.fr/phytoprot/ |
| PIP | A database of potential intron polymorphism markers | http://ibi.zju.edu.cn/pgl/pip/ |
| PLACE | Plant cis-acting regulatory DNA elements | http://www.dna.affrc.go.jp/PLACE/ |
| PLANT-PIs -- | A database for protease inhibitors and their genes in higher plants | http://plantpis.ba.itb.cnr.it/ |
| PLecDom | Plant Lectin Domains server | http://www.nipgr.res.in/plecdom.html |
| PMRD | Plant MicroRNA Database | http://bioinformatics.cau.edu.cn/PMRD/ |
| PODB | The Plant Organelles Database | http://podb.nibb.ac.jp/Organellome |
| POGs/PlantRBP | A resource for comparative genomics in plants | http://cas-pogs.uoregon.edu/#/ |
| PREP Suite | Predictive RNA Editor for Plants | http://prep.unl.edu/ |
| PRGDB | Plant Resistance Genes DataBase | http://prgdb.cbm.fvg.it/ |
| PathoPlantxae | A platform for microarray expression data to analyze co-regulated genes involved in plant defense responses | http://www.pathoplant.de/ |
| Phytome | A platform for plant comparative genomics | http://www.phytome.org/ |
| Plant MPSS databases | Signature-based transcriptional resources for analyses of mRNA and small RNA | http://mpss.udel.edu/ |
| Plant snoRNA database | Search for comprehensive information on small nucleolar RNAs in plants | http://bioinf.scri.sari.ac.uk/cgi-bin/plant_snorna/home |
| PlantCARE | A database of plant cis-acting elements | http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ |
| PlantGDB | Plant Genome Database and Analysis tools | http://www.plantgdb.org/ |
| PlnTFDB | Database of Plant Transcription Factor | http://www.softberry.com/berry.phtml?topic=plantprom&group=data&subgroup=plantprom |
| PlantTribes | A gene and gene family resource for comparative genomics in plants | http://planttfdb.cbi.pku.edu.cn/ |
| PlantTFDB | Plant Transcription Factor Databases | http://fgp.huck.psu.edu/tribe.html |
| PmiRKB | Plant MicroRNA Knowledge Base - Find information about plant microRNAs | http://bis.zju.edu.cn/pmirkb/ |
| SALAD | Surveyed contained motif ALignment diagram and the Associating Dendrogram | http://salad.dna.affrc.go.jp/salad/en/ |
| The Adaptive Evolution Database (TAED) | A phylogeny based comparative genomics tools | http://www.bioinfo.no/tools/TAED |
| The Plant DNA C-values Database | Search for information on plant DNA C-values and genome sizes. | http://data.kew.org/cvalues/homepage.html |
| The PlantsP Functional Genomics Database | Search for information on plant kinases and phosphatases | http://plantsp.sdsc.edu/ |
| The PlantsT Functional Genomics Databases | Search for genes and proteins involved in plant membrane transportation | http://plantst.sdsc.edu/ |
| The TIGR Plant Repeat Databases | A Collective Resource for the Identification of Repetitive Sequences in Plant | http://www.tigr.org/tdb/e2k1/plant.repeats/index.shtml |
| The TIGR Plant Transcript Assemblies database | Search for plant EST and cDNA sequences from this comprehensive collection | http://plantta.tigr.org/ |
| TropGENE-DB | A Multi-Tropical Crop Information System | http://tropgenedb.cirad.fr/ |
| UK CropNet | A collection of databases and bioinformatics resources for crop plant genomics | http://ukcrop.net/ |
| openSputnik x97 | A database for annotated expressed sequences tags information and comparative plant genomics analysis | http://sputnik.btk.fi/ |
Plant studies involving the analysis of biological macromolecules are collectively termed plant omics. Omics is a broad field of study encompassing subfields like genomics, transcriptomics, proteomics, metabolomics, phenomics, glycomics, lipidomic, etc. Plant genomics involves studying the compositions, organizations, functions, and structures of genetic materials (DNA/RNA) and molecular genetic networks of interactions in the plant genome.17,18 While genome structure and organization are studied in structural genomics,13,19 functional genomics investigates the functions, interaction, and regulation of gene and gene products.5,20
Plant functional genomics is a goldmine in agronomic traits improvement. It incorporates other omics approaches like transcriptomics, proteomics, metabolomics, phenomics, etc.16,21 (Figure 3). Other aspects of genomics are epigenomics, mutagenomics, and pangenomics.22 Epigenetic changes, such as histone modifications, small RNA and DNA methylations occurring at the genomic phase, are dissected within epigenomics. Mutagenomics is used to explore mutation events mediating modified genotype and phenotype in mutant species. Pangenomics studies the whole set of genomic sequences present in the entire population of a species. It also explores dispensable genomes that are individual specific or partially shared. Mutagenomics and pangenomics are new omics techniques in crop sciences.22,23
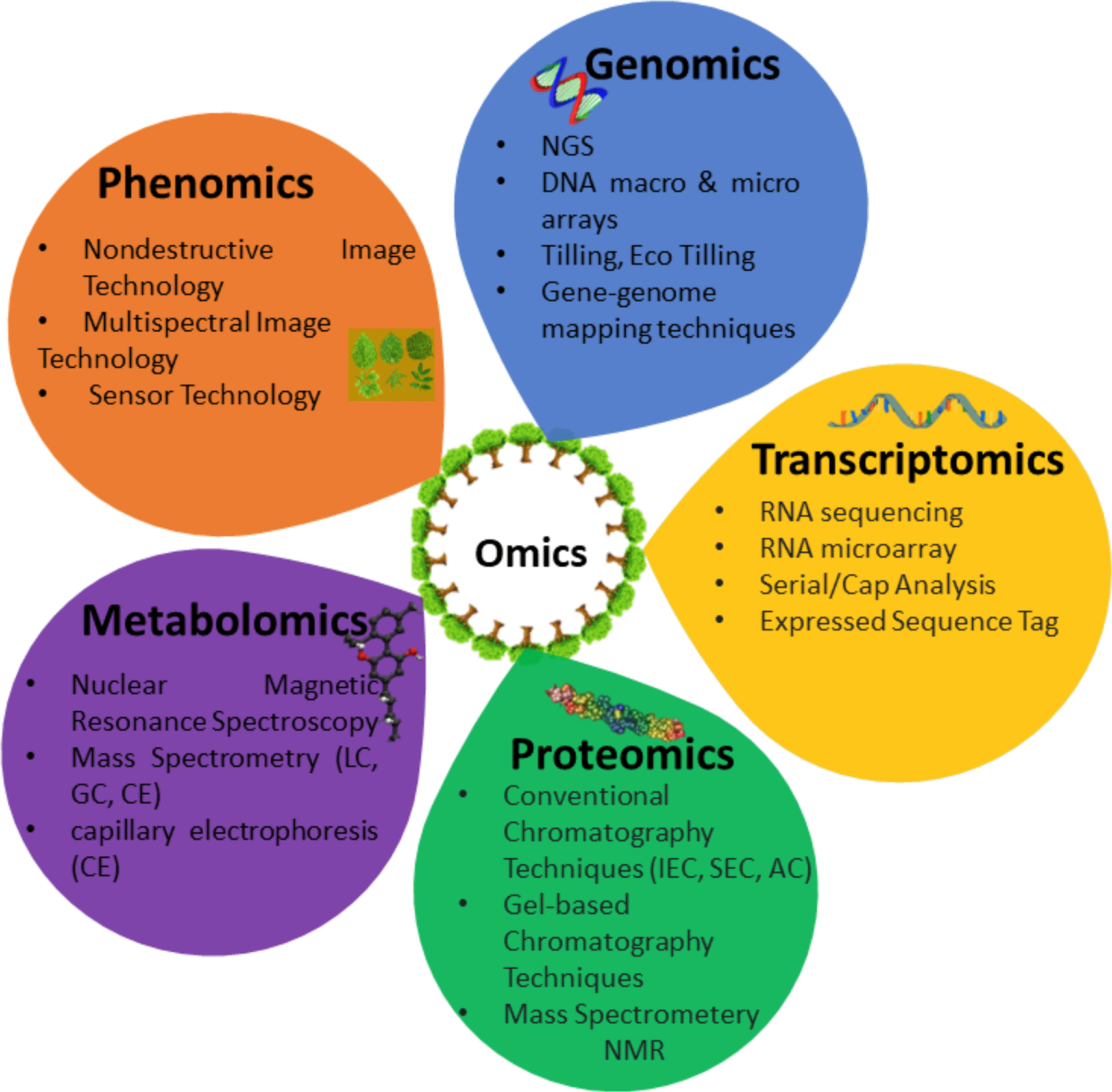
Representation of major omics approaches in plant molecular studies and the methods utilized in conventional analysis of plant omics datasets.
Plant transcriptomics involves investigating the control of plant metabolite production processes at the RNA level. Transcript-level gene expression control regulates the whole plant's development and growth.24 Plant proteomics explores the structural and functional features of proteins in a living organism. It encompasses studies on plants’ typical morphological and physiological properties.16,22 The role of proteins in controlling the plant metabolic processes is also studied in plant proteomics, especially in medicinal plants.25
Plant metabolomics involves profiling primary and secondary metabolites in plants.26 Metabolic data are useful when developing metabolic correlation networks. These networks can aid comparative analysis of cellular compartments such as carbon and nitrogen transport and partitioning in plants.27 In addition, the molecular and cellular regulation of different enzymatic processes can also be investigated.25 Plant phenomics involves the systematic study of phenotypes such as plant composition, growth, and production analysis. This study can be conducted both in controlled environments and in the field. Field phenomics involves the measurement of phenotypes that exist under both cultivated and natural conditions. Studies in controlled environments involve glasshouses, growth chambers, and other systems where growth conditions can be manipulated.28 These multi-omics approaches have emerged successful for plant research, including agronomic traits improvement over the last few decades. Agronomic traits are desirable plants’ genetic or phenotypic features, i.e., quality traits, disease resistance, pest resistance, insecticide tolerance, temperature, drought, and other adverse environmental factors tolerance traits. Quality traits encompass morphological features like plant height, seed weight; physiological features like chlorophyll content and photosynthetic rate29; economic features like improved crop yield, processing, and storage; pharmaceutical and industrial features like the elimination of toxins and allergen, increased nutritional or dietary value and increased medicinal values.30 Recent research suggests that when multi-omics technologies are integrated, they can be better harnessed to improve genetic development, crop breeding science, plant stress resistance, and other agronomic traits.22,23
Genomics-assisted pre-breeding is a genetic manipulation strategy to improve agronomic traits of interest in plants at the DNA level.31 Genomics-assisted pre-breeding approaches positively contribute to the efficiency of diseases and climate-resilient crop development.32–34 Crop breeding across the globe has relied on a series of phenotypic selection and crossing before the genomic era to generate superior crop genotypes.35 Genome sequence availability has paved the way for identifying all genes and genetic variants associated with agronomics traits.36,37 Besides, it has made it possible to assess genotype level changes incurred during breeding processes.38 Plant breeders have utilized genomics and bioinformatics in gene-level resolution of agronomic variation using quantitative trait loci (QTL) mapping39–41 and genome-wide association studies (GWAS).42,43 For instance, studies have recently been conducted to develop multiple stress-adaptable rice species that are disease and climate resilient using genomics-assisted breeding techniques such as quantitative trait locus (QTL), gene/markers-phenotype association and phenotype selection.44–46 Pea breeding projects used genetic marker-trait associations to boost valued yield and market-preferred agronomic traits.47,48 Miedaner et al.43 used high-density genotype arrays and comprehensive phenotyping of the same species population across diverse conditions, locations, and seasons in genomic selection and population mapping to speed the breeding of disease resistance traits in maize, small-grain cereals, and wheat. Hu et al.39 harnessed genomic selection (GS) and genome-wide genetic variants to prevent reiterated phenotyping in breeding cycles. These studies indicate new breeding techniques such as speed breeding and genomic selection to boost genetic and trait improvements. However, the lack of robust phenotypic data limits the efficient utilization of available genomic information and technologies in genomics-assisted breeding.
Variation in gene content among individuals within the same species is caused by genetic variation ranging from single-nucleotide polymorphisms to substantial structural variants (SVs). Due to human and natural selection acts, this variation offers the raw material on which evolution occurs.49,50 Deviation in agricultural plants’ phenotypic and genetic characteristics is referred to as crop diversity.36 The understanding of crop diversity is enhanced by plant genomics at both species and gene levels.51 According to recent research, a single reference genome is insufficient to capture a species' entire genetic diversity landscape. Pan-genome analysis provides a platform for evaluating a species' genetic diversity by looking at its whole genomic repertoire. Pan-genomic studies have shed new light on the landscape of diversity and improvement of major crops such as Brachypodium distachyon,52 Brassica Spp.,53–55 maize,56 rice,57,58 soybean,59 wheat60 etc. Evolution in plant diversity is correlated with and relatively predictable by heterogenous biotic and abiotic environmental stress induced by global climate change. These stresses, in turn, affect crop yield and crop-growing seasons.61 A study on natural plant populations shows that the organization and evolution of plant populations’ diversity at all genomic regions is nonrandom at the molecular and organismal level.62,63 Therefore, plants can evolve under climatic gradients resulting in clinal adaptation. Hence, the breeding of climate resilience crops can be facilitated by understanding the genomic basis of clinal adaption in crop species.64
Biotic stresses are instigated by living organisms such as insects, parasitic plant nematodes, diseases, or weeds in production agriculture.65 Genomics approaches to biotic stress include ribonucleic acid interference (RNAi) silencing and transgenesis. RNA interference (RNAi) silencing is employed against viruses and some fungi, while transgenesis has been exploited to develop resistance against some fungi, for example, Fusarium head blight.66 Genome-wide identification and expression analysis in legume crops also revealed the role of small RNA biogenesis mediators in biotic stress response regulation.42
Abiotic stresses, such as low or high temperatures, heavy metals, insufficient or excessive water, high salinity, ultraviolet radiation, are hostile to developing plants, resulting in significant wane in crop yield worldwide.67 According to Kumar et al.,28 knowledge of plants' response to abiotic stresses can be enhanced by integrating information generated from metabolomics and proteomics with genomics data. Sustenance of yield in crops threatened by abiotic stresses is a significant challenge in breeding resilient crop varieties.68 A study on Brassica oleracea shows that heat stress transcription factors are integral to signal transduction pathways functioning in response to environmental stresses and are suggested to contribute significantly to various stress responses.62 Heat stress transcription factors genes were identified in the in silico analysis of B. oleracea. The identified genes may be exploited in developing crop varieties resilient to global climate change.1,62
Population genomics is employed to study adaptation and speciation. Population genomics datasets are used in GWAS to detect the genes responsible for adaptive phenotypic variations of large plant population samples.35,69 For instance, Bamba et al. identified specific adaptation loci in a GWA study and unveiled the molecular basis of genetic trade-offs. It also showed that ecological fitness could be predicted by polygenic effects of several loci associated with local climate.35 Medicinal plants’ diversity is of exceptional interest because of their ethnomedicine role. GWAS studies on adaptive genotypic and phenotypic variation provide a framework to assess the diversity of medicinal plant application across different cultures and infer modifications in plant use over time.70,71 Other genomic approaches such as genomic selection, nested association mapping, genetic diversity, and allele mining have been integrated into crop improvement programs to address the genetic issues associated with maize productivity and nutritional contents.72
Plant omics studies have greatly helped our understanding and interpretation of plant responses to ecological influences and their contribution to key developmental processes important for crop yield and food quality. However, there are still some problems, such as a lack of data integration and robust techniques for phenotype-genotype correlation. Also, the use of omics-derived knowledge and tools in plant improvement strategies is limited and difficult. As a result, there is a pressing need to promote the integration of computational biology and plant genomics to benefit plant improvement.6,7
Machine learning (ML) is a computer science field that utilizes algorithms to learn and capture the characteristics of target patterns of complex datasets.73 Machine learning algorithms are generally classified into the following categories; supervised, semi-supervised, unsupervised, reinforcement, and deep learning.74 A supervised ML algorithm is trained using a labeled dataset. It learns to respond more accurately based on these training sets by comparing its output with the given input.67,73 Semi-supervised algorithms provide a tool that harnesses the potential of both supervised and unsupervised learning. These algorithms are ideally adapted for model building and can be used for classification, regression, and prediction.75 Unsupervised learning is all about identifying unexplained existing patterns from the data to generate pattern rules. Unsupervised learning is a learning approach focused on statistics and thus applied to the issue of discovering a hidden structure in unlabeled data.7,74,76
Reinforcement learning is considered an intermediate form of learning as the algorithm is only provided with an answer that tells whether the output is correct or not.75 Deep learning is built on artificial neural networks (ANN). The algorithms extract higher-level features from the raw input using multiple layers of neural networks. Learning of the algorithm can be unsupervised, semi-supervised, or supervised.73 Machine learning approaches provide unique techniques for integrating and analyzing omics data, allowing for the improvement of crops and other economically important plants. Some machine learning algorithms have been used to developed tools specifically for plant omics analysis. Table 2 highlights the existing machine learning tools for plant omics analysis. Machine learning algorithms have a broad range of applications in plant genomics. These algorithms play vital roles in genome assembly, iterative gene regulatory network inference, and the identification of true SNPs in polyploid plants.77
| Application area | Developed tools | URL | Algorithms | Selected features |
|---|---|---|---|---|
| Plants Mitochondrially Localized Proteins Prediction | MU-LOC78 | http://mu-loc.org/ | SVM and DNN | gene co-expression information, protein position weight matrix, amino acid compositions, and N-terminal sequence information, |
| plant resistance protein NBSLRR prediction | NBSPred79 | http://soilecology.biol.lu.se/nbs/ | SVM | R-protein and non-R-protein sequences attribute like sequence domain and compositional frequencies |
| Ribosomal proteins (RPs) prediction | RAMA80 | http://inctipp.bioagro.ufv.br:8080/Rama. | MLP, RF, and NB | Amino acid side chains attributes |
| Plant disease resistance proteins prediction | DRPPP81 | http://14.139.240.55/NGS/download.php | SVM, MLP, and RF | genomic sequence (satellite DNAs) |
| Geminiviruses Gene and genera classification | Fangorn Forest (F2)82 | www.geminivirus.org:8080/geminivirusdw/discoveryGeminivirus.jsp. | SVM, MLP, and RF | genomic sequence (satellite DNAs) |
| Transcriptomes for stress responses in Arabidopsis | mIDNA83 | www.plantcell.org/content/26/2/520.short#def-8 | RF with PSOL algorithm, SVM, and NN | patterns of 32 known stress-related gene expression traits and the complementary expression characteristic |
Precision breeding is a genetic engineering technique that involves reproducing organisms of the same species together to preserve desirable characteristics and create a stronger hybrid.84 Traditional statistical methods mainly used in plant breeding strategies are ineffective in plant data analysis because of the non-deterministic and nonlinear nature of plant features attributed to environment, genotype, and interaction.85 Machine learning has enabled effective plant phenotyping and data mining for patterns such as genotype and trait correlation.39 It has also been successful in genomic selection. Genomic selection is a critical method used in selecting plant species with genetic gains of interest in plant breeding. Applying different ML algorithms in building GS models has produced robust and accurate prediction.86,87 Multilayer neural networks (NNs) have been used in genetic value prediction in plant breeding. NN models are efficient in predicting genetic value, regardless of the population size, heritability, or coefficient of variation. Thus, the ANN is promising for genetic value prediction in unbiased experiments.88 Multilayer NNs have been utilized to select bean genotypes with highly stable phenotypes using 13 genotypes of common beans between 2002 and 2006. The integration of this model in plant breeding has enabled precise genetic value prediction and selection.89
Also, deep learning generated a robust prediction accuracy in grain yield compared to the conventional linear statistical methods used in traditional plant breeding when analyzing multiple traits with mixed ordinal, continuous, and binary phenotype data. Univariate and multivariate deep learning models' predictive performance was assessed using the Durum wheat (Triticum turgidum var. durum Desf.) dataset. Deep learning model performance shows that it has a promising potential to be a successful model for accurate genomic prediction in plant breeding.90 The flexibility of machine learning algorithms makes them a viable alternative to traditional parametric methods for predicting categorical and continuous responses in genomic selection.91
An ensemble of RF and SVM was implemented to improve genotype-phenotype classification using manually derived root trait datasets. The combined model accurately identified the most distinguishing root traits and corresponding cultivar differentiation. The model's performance demonstrates the potential of ML approaches in unbiased cultivars classification and trait selection.92
Additionally, predictive models have aided the integration of additive and dominance effects in GWAS and have enhanced the prediction of complex agronomic traits in polyploid plant species. For instance, a study revealed the feasibility of genome-wide prediction of potato agronomic traits despite being an autotetraploid food crop. It also shows that GWA prediction is viable in selecting breeding values in elite germplasm with substantial non-additive genetic variance.93
Plant phenomics is a systematic study of plant phenotypes.28 In recent years, plant field phenotyping has gotten a lot of attention with the possibility of crop fields' high-throughput analysis.94 The application of machine learning methods and the various technological developments for image analysis have improved quantitative crop traits assessment.92,94,95 For instance, CNN-based detection and analysis of wheat spikes using wheat field trials images captured over one planting season achieved an average accuracy of 88 to 94% across diverse groups of test images. CNN's high-performance accuracy shows that it is a robust model for genome-based selection and prediction in plant breeding.96 Also, the RF algorithm was used in plant image segmentation involving the acquisition and processing of several plant images samples.97 The predictions made by the model enabled the discovery of various parameters relevant to plant growth.94
ML learning has been exploited in identifying favorable agronomic traits, including abiotic and biotic stress resistance. ML algorithms are integrated into conventional statistical methodologies to optimize the accuracy of plants stresses prediction and detection.96 For instance, 25,000 soybean leaflets images exposed to varied diseases and nutritional perturbation were used to develop a convolutional neural network (CNN), which can infer the image features of the disease types and dietary deficiencies at high resolution. The prediction accuracy of the ML framework was very close to that of human expert diagnosis. Other plants’ induced stresses can also be identified, classified, and quantified using the model. The model can also be adapted to identify, classify, and quantify the induced stresses in other plants.95
Random forest has been used to predict metabolite and transcript markers in drought tolerance prediction using experimental drought-stressed plant field trial datasets. The low error rate recorded in the model shows that the model could be considered as an alternative model for accurate prediction and identification of molecular markers.98 RF was used to identify suitable features combination for phenotypic traits prediction using data derived from various agro-management treatment experiments. This approach achieved optimal prediction accuracy and improved plant breeding strategies by enabling maximal allocation of stress management resources.99 Sanz-Carbonell and colleagues used deep sequencing and computational approaches such as PCA and Clustering analysis to infer the biotic and abiotic stress responses regulatory network mediated by miRNA. 24 miRNAs were used in this study, all of which are known to alter expression significantly under stressful conditions. The prediction generated inference that target genes of miRNAs down-regulated under stress conditions contribute to plant response to stress, whereas miRNAs that are up-regulated control genes associated with growth and development.67 Soybean fields were screened for tolerance to soybean iron chlorosis deficiency (abiotic stress in soybean) using linear discriminant analysis (LDA) and SVM. The phenotypic data obtained from soybean fields were used in model training and predicting soybeans' iron chlorosis deficiency stress. The ML application has helped evaluate the severity of real-time stress in the soybean sector.95
Plant diseases and pests pose a significant threat to agriculture. Early identification of plant diseases and pests would aid in developing effective treatment strategies while economic losses are mitigated.100 Diverse ML approaches for precise disease recognition and prediction have been implemented in plant populations.101,102 Neural networks (NNs) have achieved impressive results in plant disease prediction using image classification. A deep convolutional network was implemented in leaf image classification model for disease recognition. The developed model showed a high predictive performance in distinguishing plant leaves from their surroundings and recognized 13 plant diseases types on healthy leaves.103 In another approach, a heterogeneous ensemble of deep-learning-based neural network models was used in detecting tomato plants diseases and pests using images collected on-site by imaging devices of varying resolutions. The ensemble model successfully handled image complexity in the plant's surrounding area and recognized nine different diseases and pests.100,104 Therefore, deep CNNs are promising in automatic classification and detection of diseases traits from leaf images. In addition, CNN has shown optimal performance when implemented in plant–pathogen interaction, pest, and disease recognition in some studies. These studies include prediction of pests and diseases occurrence in cotton105; rice plant diseases and pests recognition106; rice blast disease prediction107,108; image-based potato tuber disease detection109 and so on. The CNN model is a high-performing method for detecting plant diseases, and it can be implemented and optimized for practical applications.
SVM has also been used for weather-based rice blast prediction and has proven suitable for plant disease forecasting with incredible predictive accuracy. A world-first SVM-based web server for rice blast prediction was developed. Plant scientists and farmers have benefited from this tool, especially in their decision-making.110 In the pixel-wise quantification and identification of powdery mildew diseased barley tissue, SVM classification was used to establish hypersensitive response spots using multispectral imaging of diseased barley plants. SVM application enabled precise automatic identification of barley interaction with powdery mildew.111
Recently, a data-driven ML approach named ApoplastP was proposed. RF classifier is the base algorithm for ApoplastP and has shown high performance in predicting protein localization in plant apoplast. At first, differences in the constituents of apoplastic and intracellular plant proteins were unknown. However, the advent of ApoplastP enabled the exploration of differences in the composition of plant proteins. The plant apoplast is integral to plant–pathogen interactions, transport, and intercellular signaling. Hence, integrating and optimizing machine learning algorithms in apoplastic localization prediction will aid functional studies and help predict whether an effector will localize to the apoplast or enter the plant cells.112
Also, RF has been implemented to build an inter-species protein–protein interaction (PPI) prediction model using Arabidopsis–pathogen PPI data acquired both experimentally and from PPI public databases-UniProt. A critical assessment of the model performance showed that random forest integration with linear statistical methods using sequence information and network attributes as model features resulted in substantial and robust improvement in performance.24
In addition, RF classification has been used to exploit protein biomarkers' potential in precision breeding using biomarkers generated assays of 104 potatoes (Solanum tuberosum) peptides. These peptides were selected using diagonal linear discriminant analysis, bagging, principal component analysis (PCA), and SVM and then classified with RF classifiers. The ML algorithms' application helped identify Phytophthora infestans resistance in leaves, tubers and its effect on plant yield using potato leaf secretome data.109 Early disease detection enables farmers to use timely and targeted crop protection strategies. With the use of ML, researchers have improved the accuracy of object detection and recognition systems dramatically.
ML applications in plant genomics and agronomics have majorly contributed to efficient breeding of crops with desirable agronomic traits, plant phenotyping, genetic trait prediction, and precise disease prediction such as in rice, soybeans, maize, beans, etc.72,95,106,110 However, several limitations still exist. Firstly, the black-box nature of some sophisticated ML algorithms inhibits interpretation. The plant research community is more interested and fascinated with the biological implications of the prediction than the accuracy of the predictive model. Hence, there is a need for further processing and careful interpretation of the predictive model output using conforming biological knowledge. Additionally, the dimensionality of omics datasets poses challenges such as multicollinearity, overfitting, and sparsity which are difficult to avoid. Though contemporary machine learning methods and the huge sample size can partially alleviate these problems, the model’s accuracy can be significantly enhanced by using different fine-tuning, augmentation, and optimization techniques.107 Data integration from various sources is necessary for GS-assisted breeding and other trait improvement approaches.113 Simultaneous analysis of multiple omics datasets can advance our understanding of complex biological phenomena.78,79 Another challenge is the limited and inconsistent information on plant-pathogen interaction phenotypic information. The ML models used in plant disease recognition can be extended by enriching the plant disease database with plant-pathogen interaction phenotype data. Developing more robust classification algorithms with an expanded number of diseases classes will improve plant disease recognition and forecasting.103,105,108,106 Finally, a comprehensive plant database must be constructed to facilitate comparative studies and promote research collaborations on critical plant science problems.
Machine learning has shown tremendous promise in studying enormous high-dimensional data sets, although it is still limited in plant molecular studies application. An in-depth understanding of ML models will stimulate ML implementation for plant biological data analysis. As sequenced plant genome data continues to accumulate, ML will accelerate all plant genomic research fields, including identifying genes associated with biotic and abiotic stress resistance and other genes with significant functions, understanding gene regulation mechanisms, exploring plant genome genetic framework, and estimating breeding values. These advancements would help agricultural researchers improve the quality and yield of crops with stronger tolerance to abiotic and biotic stress and other plant health-threatening issues.
OSF: Extended data for “Machine learning algorithms: their applications in plant omics and agronomic traits’ improvement”, https://doi.org/10.17605/OSF.IO/TE6GC.14
Files included:
Supplementary Table 1. Published sequenced plant genomes. Hundreds of plant genomes have been sequenced and published since 2000. The statistics for each genome are taken from the publication, despite several model plants having significant updates to genome assemblies and gene counts. NA, data not available in publication; Mb, megabases; kb, kilobases.
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the topic of the review discussed comprehensively in the context of the current literature?
No
Are all factual statements correct and adequately supported by citations?
Yes
Is the review written in accessible language?
Yes
Are the conclusions drawn appropriate in the context of the current research literature?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bioinformatics, Quantitative Genetics, Statistics, Plant Breeding
Is the topic of the review discussed comprehensively in the context of the current literature?
No
Are all factual statements correct and adequately supported by citations?
No
Is the review written in accessible language?
Yes
Are the conclusions drawn appropriate in the context of the current research literature?
No
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Artificial intelligence in genetics, Systems biology modelling, Quantitative genetics modelling, Multi-omics big data analysis
Is the topic of the review discussed comprehensively in the context of the current literature?
No
Are all factual statements correct and adequately supported by citations?
No
Is the review written in accessible language?
Yes
Are the conclusions drawn appropriate in the context of the current research literature?
No
References
1. Washburn JD, Mejia-Guerra MK, Ramstein G, Kremling KA, et al.: Evolutionarily informed deep learning methods for predicting relative transcript abundance from DNA sequence.Proc Natl Acad Sci U S A. 2019; 116 (12): 5542-5549 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Machin learning, plant systems biology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 04 Nov 22 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)