Keywords
Corbicula fluminea; mitochondrial genome; phylogenetic analysis
This article is included in the Genomics and Genetics gateway.
Corbicula fluminea; mitochondrial genome; phylogenetic analysis
Clams of the genus Corbicula are currently spread worldwide, and they cause great ecological threats and tremendous impacts in the ecological system (Counts 1981; Gomes et al. 2016; Peñarrubia et al. 2017; Hünicken et al. 2019; Douglass et al. 2020). However, their taxonomic status and systematic status are still unclear (Wang et al. 2018; Haponski 2019; Bodon et al. 2020; Ramli et al. 2020). Corbicula fluminea is an important component of Asian freshwater macrobiota and has always been consumed as food in East Asia (Wang et al. 2014; López-Soriano et al. 2018; Zhang et al. 2019; Sano et al. 2020). In this work, the complete mitochondrial genome of Corbicula fluminea from Dongting Lake was sequenced, and the phylogenetic relationships among Venerida were investigated. These results could contribute to distinguishing the taxonomic placement and systematic status of genus Corbicula in further studies.
The specimens (Corbicula fluminea) used in this work was collected from Dongting Lake (geographic location: 28°46′45″N, 112°41′22″E), in Hunan province, China in August 2018.
After sampling, the living specimens were stored in College of Animal Science and Technology, Hunan Agricultural University, the voucher number is DT1808-118.
To obtain the total genomic DNA, the 40mg frozen muscle of the foot was derived from the specimen. DNA was obtained by DNeasy™ Blood & Tissue extraction kit (Qiagen, Hilden, Germany). The library was conducted by Berry Genomics Co. Ltd (Beijing, China) according to the Illumina TruSeq Nano DNA library Prep Kit and the manufacturer’s recommendations. DNA fragments were selectively enriched using Illumina PCR Primer (F: 5′-AATGATACGGCGACCACCGAGA-3′ and R: 5′-CAAGCAGAAGACGGCATACGAGT-3′) Cocktail in a PCR reaction with the following cycling conditions: 95°C for 3 minutes, followed by 8 cycles of 98°C for 20 seconds, 60°C for 15 seconds and 72°C for 30 seconds, with a final extension step at 72°C for 5 minutes. The raw reads of the mitogenome sequence were obtained from the platform of Illumina HiSeq 2500. The adaptors and low sequencing qualities reads (N bases exceeding 10% and more than 50% of phred quality score ≤ 5) were trimmed and reduced using the program Trimmomatic v 0.38.0 (Bolger et al. 2014).
The sequence reads of untrimmed high-quality were assembled by the program MITObim (Hahn et al. 2013). The annotation was performed using MITOS WebServer (Bernt et al. 2013) for the entire mitogenome of Corbicula fluminea from Dongting Lake, and adjusted manually in Geneious Prime v2020.2.2 based on the published mitogenomes of Venerida species.
To validate our data, the CDS sequences of 19 Venerida species (GenBank accession numbers are shown in Figure 2) were aligned. The phylogenetic analysis among Venerida was investigated using maximum-likelihood (ML) and Bayesian inference (BI) approaches. The ML tree was estimated by RAxML version 8.2.12 (Stamatakis 2014) and the BI tree was estimated by MrBayes version 3.2.7 (Ronquist et al. 2012), with GTR-GAMMA model. RAxML was analyzed with 1,000 bootstrap replicates (-m GTRGAMMAI -f a -x 1 -N 1 -p 1 -N 1000), 1 million generations Markov chain Monte Carlo iterations (nCat=4) were analysed and every 1,000 generations were sampled with the initial 10% of samples removed as “burn-in” in MrBayes. The average standard deviation of split frequencies (<0.01) was used to assess the convergence.
A phylogenetic relationship was estimated based on 13 protein-coding genes sequences from the mitogenome sequences of Corbicula fluminea from Dongting Lake and other 19 Venerida species (Figure 2). The study has provided a complete mitochondrial genome of Corbicula fluminea from Dongting Lake. As a phylogenetic tool, it could contribute to systematic and comparative analysis for resolving evolutionary relationships.
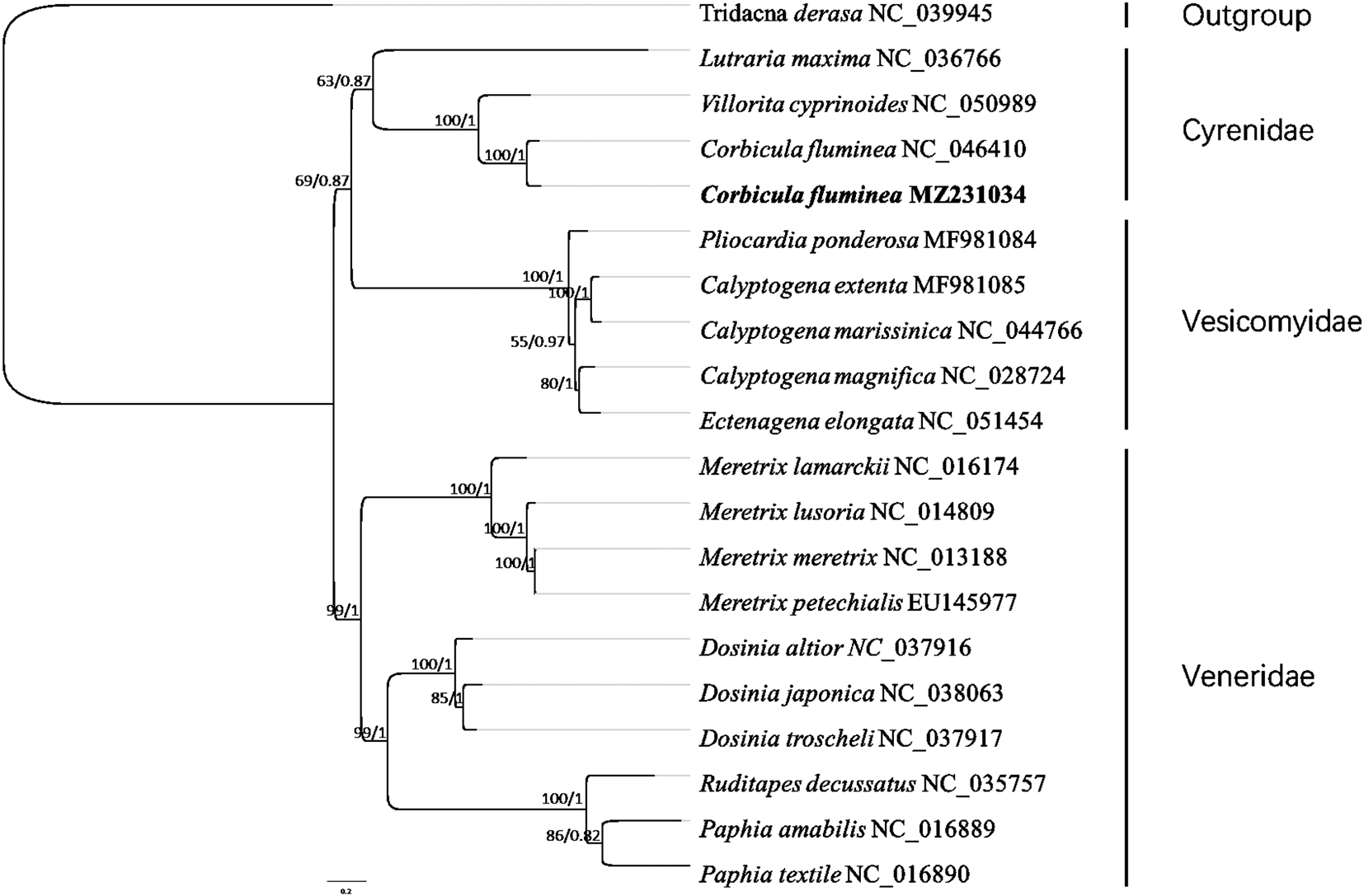
The nodal numbers indicate the bootstrap support values (left) and the posterior probability (right). Genbank accession numbers for the sequences are indicated next to the species names.
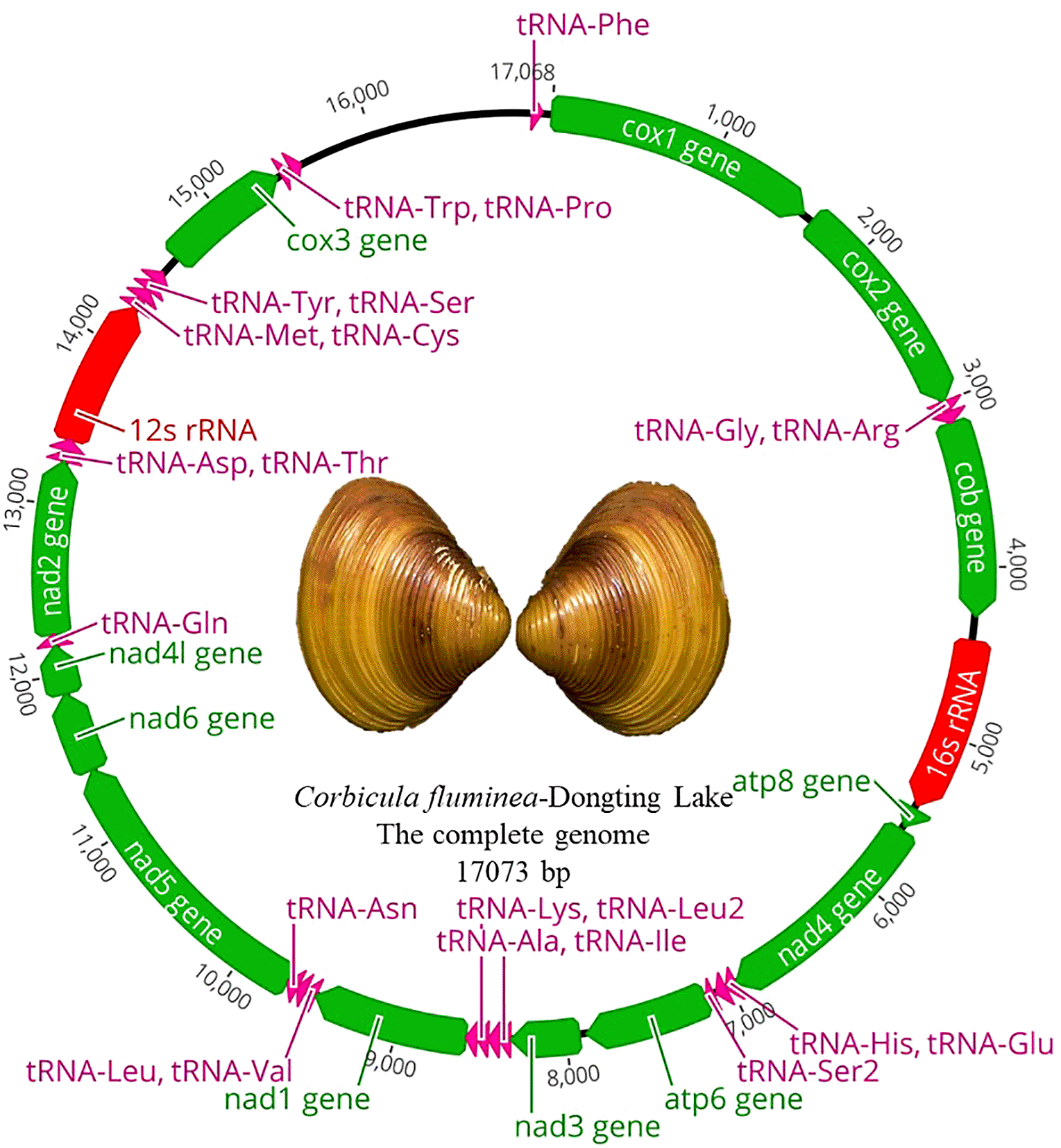
The complete circular mitogenome of Corbicula fluminea from Dongting Lake (GenBank accession no. MZ231034.1) is 17,073 bp in total length. As other Venerida mitogenomes, 37 mitochondrial structural genes were found in the mitogenome of Corbicula fluminea from Dongting Lake, including 13 protein-coding genes, two ribosomal RNA genes (12S and 16S rRNA) and 22 transfer RNA genes (Figure 1). Both are encoded on the positive strand.
NCBI Gene: Complete mitochondrial genome of Corbicula sp. DT118, SRA
Accession number: SRR14692229
Accession number URL: https://www.ncbi.nlm.nih.gov/sra/SRR14692229
NCBI Gene: Corbicula sp. QL-2021 mitochondrion, complete genome
Accession number: MZ231034.1
Accession number URL: https://www.ncbi.nlm.nih.gov/nuccore/MZ231034.1
All the clams and experimental protocols involving experiments of this study were approved by the Animal Ethics Committee of Hunan Agricultural University, Hunan, China.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Are the rationale for sequencing the genome and the species significance clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Mollusc physiology and molecular biology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 24 Feb 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)