Keywords
COVID-19, pandemic, SARS-CoV-2, Omicron variant, emerging disease, global health, virus, genome, mutation
This article is included in the Pathogens gateway.
This article is included in the Emerging Diseases and Outbreaks gateway.
This article is included in the Coronavirus (COVID-19) collection.
COVID-19, pandemic, SARS-CoV-2, Omicron variant, emerging disease, global health, virus, genome, mutation
This revision was made in response to the outstanding and critical comments from Dr. Wang and Dr. Revindran-Stam. Among other things, we have added the phylogenetics of the Omicron variants, discussion about the sublineage of the Omicron variant (BA.1, BA.2, BA.3, BA.4, BA.5 and XE), added the mutations outside the S-proteins (e.g., ORF and NSP) and provided figures depicting the mutation sites, clarify the usefulness of Ct in the Omicron infection, as well as added a paragraph discussing the clinical presentations of the Omicron variant as compared to other COVID-19 variants. We believe that this revised manuscript has answered most (if not all) of the concerns raised by the esteemed Referees.
See the authors' detailed response to the review by Leyi Wang and Vanessa Revindran-Stam
Coronavirus disease 2019 (COVID-19) has been a major cause of morbidity and mortality worldwide since December 2019. Up to July 2022, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected more than 570 million people and contributed to the death of more than 6.4 million individuals around the world. Intriguingly, after more than 2 years of its existence, the infection rate remains high, and the pandemic has not been resolved. Following the devastating impacts of the B.1.617.2 (Delta) variant, predominantly in health and socioeconomic sectors, there was a high expectation that the viral disease could be tamed by the rapid, collaborative and evolutionary development of COVID-19 vaccines. Indeed, the massive vaccination program in several countries has successfully reduced the fatality of the disease and together with the implementation of strict public health and social control measures (PHSCM), the infection rate could also be lowered.1–4 For example, in the United States (US), COVID-19 vaccines reduced the overall attack rate (i.e., number of new cases during specified time interval divided by the total population at start of time interval) in vaccinated individuals by 4.4% on day 300 from the start of vaccination (9% in the unvaccinated group vs. 4.6% in the vaccinated group), as well as the rate of hospitalization, intensive care unit (ICU) occupancy, incidence of major adverse events and mortality.1 Despite the reported effectivity decline of BNT162b2 and ChAdOx1 vaccines against the Delta variant (10–13% and 16% lower than B.1.1.7 (Alpha) variant, respectively),5 vaccination was proven to remain effective in reducing infection and accelerating viral clearance,2 as well as lowering mortality caused by the Delta variant.4
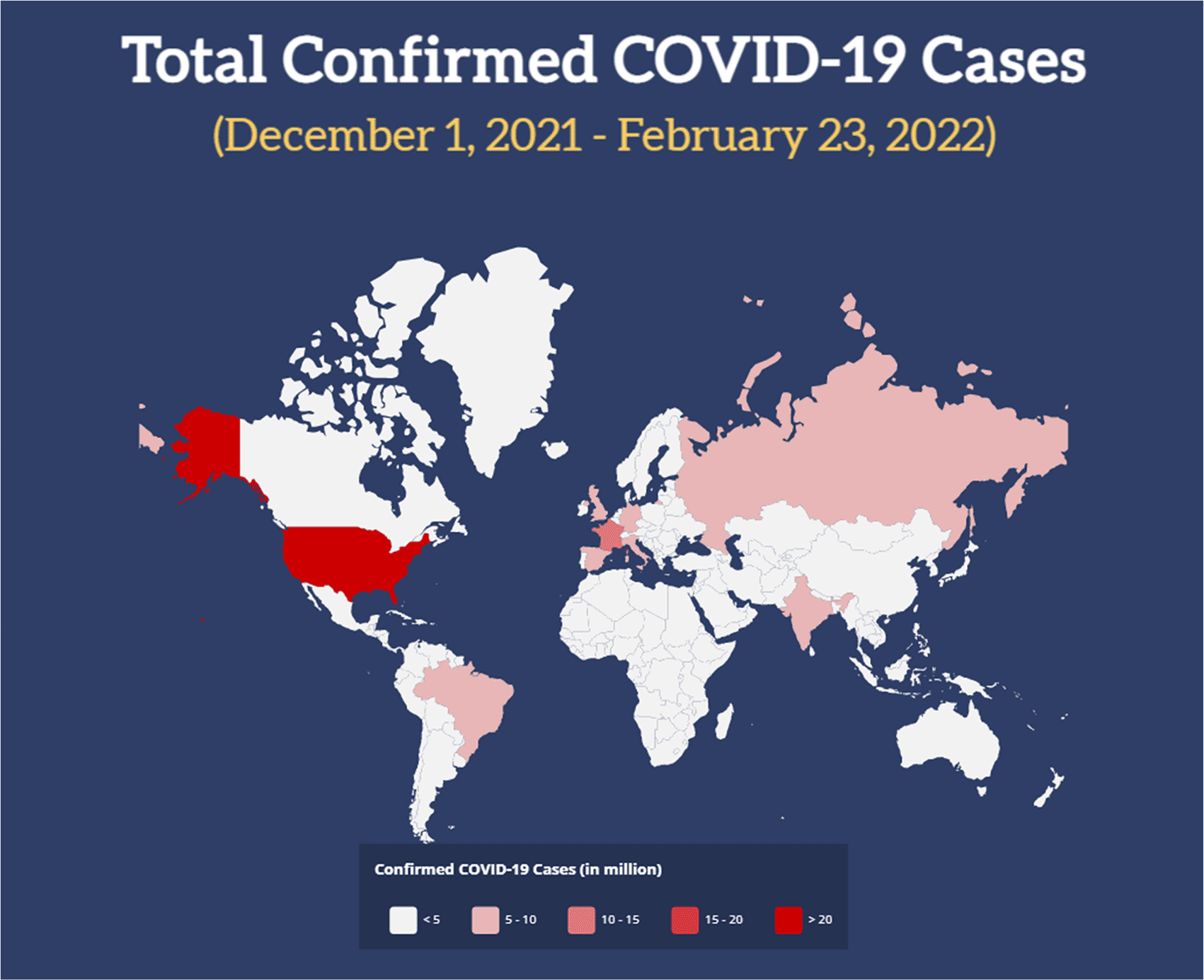
However, a glimpse of hope to end the pandemic was once again challenged by the presence of the new COVID-19 B.1.1.529 (a.k.a. Omicron) variant. Since its first reported appearance in South Africa in November 2021, it has rapidly taken the attention of experts around the world. A few days after its appearance, the World Health Organization (WHO) immediately classified Omicron as a Variant of Concern (VoC). Since then, it has vastly spread from Africa to Europe, then Asia, Australia and America (Figure 1). Since early January 2022, the Omicron variant has become the major COVID-19 variant in most countries (Table 1)6,7 and contributed to the rise of COVID-19 incidence from ~600,000 new cases in late November 2021 to ~3.5 million cases in late January 2022.
Phylogenetics of the Omicron variant
Several studies have attempted to investigate whether there are evolutionary connections between the Omicron variant and its predecessors.8,9 For instance, using the ultrametric and metric clustering method, Kandeel et al.8 found that the Omicron variant was not closely connected with the previous SARS-CoV-2 variants, but rather formed a new monophyletic clade on its own. Similarly, another phylogenetics study by Sun et al.9 discovered that the Omicron variant was not evolutionarily derived from the Delta variant and did not share a common mutation profile with it. Of note, the Omicron variant first appeared when the Delta variant dominated the global COVID-19 infection back in late 2021, so it was hypothesized that the Delta variant evolved into the Omicron variant. Instead, they found some similarities between the Omicron and the Gamma variants,9 as also reported by Kannan et al.10 Using a different method for phylogenetics analysis (i.e., the Neighbor-joining method), Kandeel et al. revealed a connection between the Omicron variant and the Alpha variant. Furthermore, among the circulating SARS-CoV-2 variants, they found that the Alpha variant had the least nucleotide changes as compared to the Omicron variant, highlighting the close connection between the two.8
A study conducted based upon the genome sequencing data of 108 samples collected from patients infected with the Omicron variant11 revealed that this variant possessed 61-64 mutations, 54 of which were single nucleotide polymorphisms (SNPs). Of those, 34 mutations were positioned at the spike (S) proteins (Figure 2) and 32 of those spike mutations were non-synonymous, which means that the mutations alter the amino acid sequences.11 Importantly, the mutations in the Omicron variant were reported to be present in all three key regions in the spike of SARS-CoV-2: The receptor binding domain (RBD), N-terminal domain (NTD) and furin cleavage site (FCS). Overall, 15 of the spike mutations were located in the RBD, a region that is responsible for the viral attachment to the cell surface (Table 2).
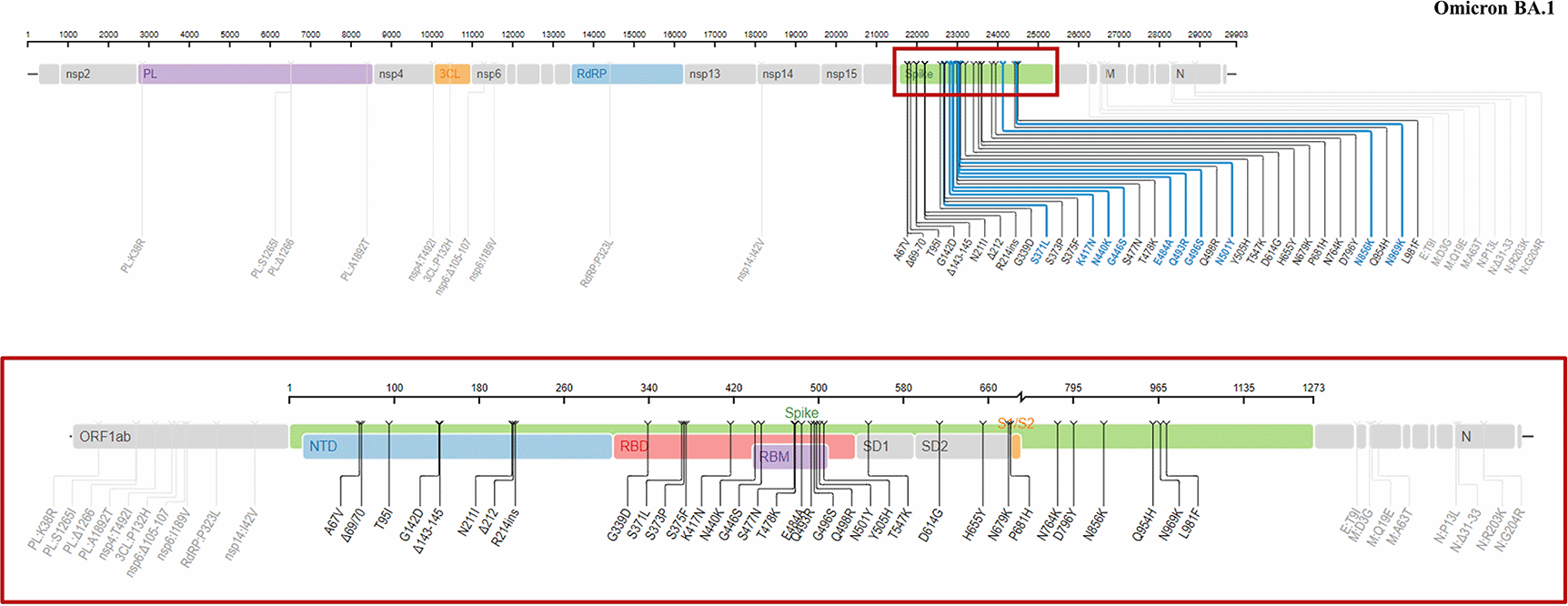
The red box depicts the mutations located in the S-proteins of the virus. In total, there are 34 mutations in the spike protein and the rest are located outside of this region (e.g., in the non-structural protein (NSP) or the open reading frame (ORF) regions). This figure was obtained from the Stanford University Coronavirus Antiviral and Resistance Database under license CC BY-SA 4.0.
| NTD | RBD | FCS & adjacent |
|---|---|---|
| A67Va,b | G339Da,b | T547Ka,b |
| IHV68Ia | S371La,b | D614Ga,b |
| del69-70b | S373Pa,b | H655Ya,b |
| T95Ia,b | S375Fa,b | N679Ka,b |
| GVYY142Da | K417Na,b | P681Ha,b |
| del142-144b | N440Ka,b | N764Ka,b |
| Y145Db | G446Sa,b | D796Ya,b |
| NL211Ia | S477Na,b | N856Ka,b |
| del211b | T478Ka,b | Q954Ha,b |
| L212Ib | E484Aa,b | N969Ka,b |
| Ins214EPEb | Q493Ra,b | L981Fa,b |
| G496Sa,b | ||
| Q498Ra,b | ||
| N501Ya,b | ||
| Y505Ha,b |
a was reported by Ma et al.11 and
b was reported by the United States Centers for Disease Control and Prevention (CDC). (FCS = furin cleavage site; NTD = N-terminal domain; RBD = receptor binding domain).
Mutations in RBD
As displayed in Table 2, the N501Y mutation that converts amino acid asparagine to tyrosine at position 501 was detected in the Omicron variant. Of note, the N501Y mutation was also identified in other COVID-19 variants (e.g., C.1.2, Alpha, Beta and Gamma).12 This particular mutation enhanced the binding affinity of the RBD to angiotensin converting enzyme type-2 (ACE2) receptor in the surface of the host cell.13 As a consequence, the Omicron variant could have a stronger attachment to the host cells than some of the other COVID-19 variants, fostering its transmissibility. Meanwhile, the mutation Q498R (converting glutamine to arginine at position 498) of the RBD alone negatively affected the protein stability and binding. However, due to its known epistatic effect with N501Y, the combination of both mutations was shown to increase the affinity of the RBD to ACE2 receptor by 4-fold.14 In contrast with the N501Y mutation, the Q498R mutation has not been seen in other COVID-19 variants. In addition to their effect on the binding affinity to ACE2 receptor, Omicron-associated mutations in the RBD could also promote the escape from existing neutralizing antibodies. In the study done by Cao et al.,15 the effect of Omicron-associated mutations in the RBD region on neutralizing antibodies was assessed using a high throughput yeast display screening. As the result, K417N, G446S, E484A and Q493R mutations assisted the virus to evade neutralizing antibodies, especially the ones which epitope overlapped with the ACE2 binding motif (epitope group A-D). Also, there was evidence that G339D, S371L and N440K mutations of the RBD could facilitate the virus evasion from other neutralizing antibodies (epitope group E-F). Overall, of the 247 antibodies tested, 85% were evaded by Omicron, suggesting the potentially low efficacy of preexisting neutralizing antibodies against the Omicron variant.15
Mutations in NTD
Several mutations in the NTD region were shown to have significant effect on viral infectivity and the modulation of immune evasion. For example, the del69-70 boosted the infectivity of the Alpha variant through the elevation of cleaved spike incorporation into virions.16 Conversely, T95I mutation that was previously identified in the B.1.617.1 variant seems not to be substantially involved in immune evasion due to its location outside of the antigenic supersite.17 Interestingly, two cases of BNT162b2 and mRNA-1273 vaccines breakthrough infection were reported in COVID-19 patients harboring T95I mutation of the NTD, as well as the del142-144 mutation of the NTD and D614G mutation of the FCS, indicating the possible contribution of those mutations on viral immune escape.18 Meanwhile, evidence of a marked (4-16-fold) reduction of neutralizing capacity of COVID-19 convalescent sera against a recombinant vesicular stomatitis virus carrying SARS-CoV-2 spike protein with Y145D mutation was also reported.19
Mutations in FCS
The FCS region has been shown to be a key part of SARS-CoV-2 pathogenesis and severity. For instance, a mutant lacking FCS (ΔPRRA) displayed a reduced replication rate in a human respiratory cell line.20 Meanwhile, in regard to Omicron-associated mutations, the H655Y and N679K mutations which are located proximal to the FCS, and the P681H mutation of the FCS could enhance the SARS-CoV-2 spike cleavage and increase the transmissibility of the Omicron variant. Moreover, the P681H mutation, which was previously identified in the B.1.1.7 (Alpha) variant, was demonstrated to facilitate the viral resistance against innate immunity (i.e., interferon-β) in lung epithelial cells.21 Yet, another study on this particular mutation did not find any notable change on the spike cleavage, viral entry or intercellular spreading.22 Next, the D614G mutation, a common mutation found in existing COVID-19 variants, increased viral replication in human lung epithelia and respiratory tract by fostering the virion stability and infectivity. Moreover, it increased the viral load in the upper respiratory tract, thereby promoting viral transmission.23 However, it is important to note that the effect of such mutations on Omicron pathogenicity may differ from what was reported in previous COVID-19 variants due to distinct interactions among variant-associated mutations.
Mutations outside of the S-region
In addition to the abovementioned mutations in the spike (S) region, several mutations were detected in the open reading frame (ORF) and non-structural protein (NSP) regions of the virus (e.g., NSP3, NSP4, NSP5, NSP6, NSP12 and NSP14). Three amino-acid deletions at L3674-, S3675- and G3676- were found in ORF1a region and could play a role in altering the cells’ ability to degrade viral components. Additionally, K856R, S2083-, L2084I, A2710T, T3255I, P3395H, L3674-, S3675-, G3676-, and I3758V were also documented in ORF1a region. Meanwhile, several mutations were detected in ORF1b region, such as P314L and I1566V; while in ORF9b (i.e., a region that is deemed to be involved in the suppression of the innate immune response to viral infection), three amino-acid deletions at E27-, N28- and A29- were present together with P10S.10,24 Covariants.org also described two other mutations in nucleocapsid: R203K and G204R, which were linked with the capability to increase viral loads.
BA.1
B.1.1.529.1 is the original Omicron variant first identified in Botswana, before rapidly spreading to South Africa and all over the world. It was first reported on November 24th, 2021 by the Network for Genomics Surveillance in South Africa. As described in the previous section, this subvariant has a higher transmission rate, an increased immune evasiveness and presumably lower severity as compared to the other circulating COVID-19 variants.
BA.2
In a recent statement, WHO’s Technical Advisory Group on SARS-CoV-2 Virus Evolution (TAG-VE) has raised a concern regarding Omicron sublineage BA.2. For the past few months, globally, the proportion of BA.2 sublineage has been increasing relative to BA.1. BA.2 (B.1.1.529.2) was first identified among genome sequences submitted to GISAID database from the Philippines back in November 2021.25 Subsequently, the BA.2 sublineage was identified in viral genome sequences from more than 40 countries in January 2022, with Denmark, India, Sweden and Singapore having recorded the most cases. Early research showed that this sublineage was significantly more transmissible than the original B.1.1.529 variant and could cause more severe disease than BA.1. Moreover, it is also more resistant to neutralizing antibodies than BA.1. A study evaluating the response of neutralizing antibodies against BA.1 and BA.2 in 24 fully vaccinated individuals and 8 people with previous SARS-CoV-2 infection demonstrated that the median BA.2 neutralizing antibody titer was 1.3-1.4 times lower than the median BA.1 neutralizing antibody titer.26 Another study also discovered that BA.2 had a 1.4 times higher effective reproduction number and a higher replication efficacy in human nasal epithelial cells than the BA.1. Additionally, it was resistant to the BA.1-induced humoral immunity. Furthermore, the spike of BA.2 was more fusogenic than the one in BA.1, suggesting its higher pathogenicity than the original Omicron variant.27 Importantly, individuals with previous BA.1 infection could still contract the BA.2 sublineage and BA.2 was shown to be capable of inducing vaccine breakthrough.28
BA.2 (Figure 3) shared some genomic similarities with BA.1, with 32 similar mutations presented in both subvariants. Nonetheless, 28 distinct mutations were reported in BA.2, including 4 specific mutations in the RBD (S371F, T376A, D405N and R408S) and in the NTD (T19I, del24-26, A27S and V213G) regions.26 In addition, the absence of del69-70 mutation of NTD in BA.2 could significantly lower the identifiability of this subvariant in some PCR assays.
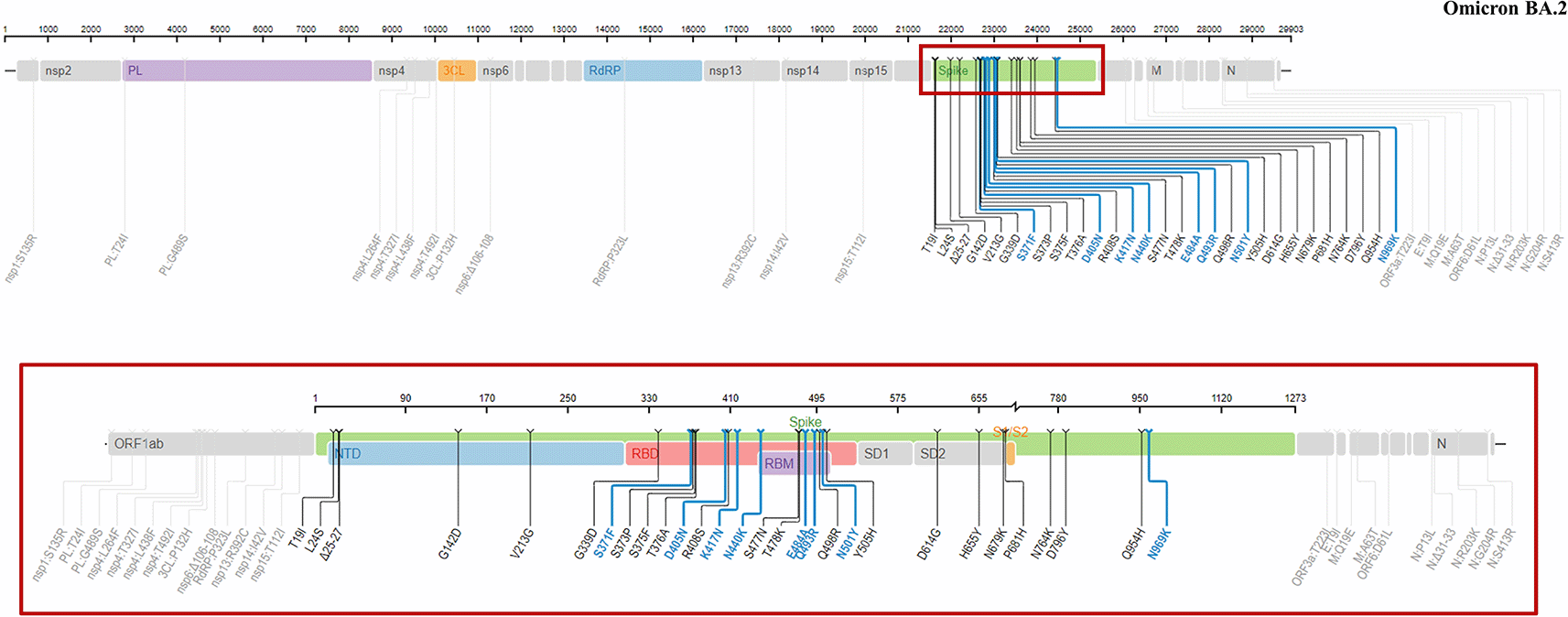
The red box depicts the mutations located in the S-proteins of the virus. This figure was obtained from the Stanford University Coronavirus Antiviral and Resistance Database under license CC BY-SA 4.0.
BA.3
BA.3 was first documented in northwestern South Africa and constituted the combination of mutations in BA.1 and BA.2. BA.3 shared most of its mutations with BA.1 and BA.2, except for one at NSP6 (A88V). It also has 15 RBD mutations, but none was distinct from that of BA.1 and BA.2.28 In the population, BA.3 caused the lowest number of cases in these three lineages. It may have been due to the loss of six mutations (ins214EPE, S371L, G496S, T547K, N856K, and L981F) from BA.1 or obtaining two mutations from BA.2 (S371F and D405N). As also found in BA.2, BA.3 retained the N501Y and Q498R mutations, which enhance the binding to ACE2 receptor, and the H655Y, N679K, and P681H mutations, which increase spike cleavage and facilitate virus transmission.29
XE
This subvariant is a hybrid or recombinant of BA.1 and BA.2, which was first discovered in the United Kingdom in January 2022. The XE subvariant has similar spike and structural proteins as BA.2, and obtains the 5′ part of its genome from BA.1. This subvariant was found to be more transmissible than BA.1 and BA.2, making it one of the most transmissible COVID-19 variants to date. Moreover, XE subvariant had a 9.8% higher growth rate than BA.2, suggesting the potentially more infectious nature of this Omicron subvariant.30 Interestingly, despite sharing some similar mutations at NSPs with BA.1 and at other sites with BA.2, XE subvariant carries 3 specific mutations that was not seen in its predecessors: NSP3 C3241T, NSP3 V1069I and NSP12 C14599T.30
BA.4 and BA.5
These two Omicron subvariants (BA.4 and BA.5) were first identified in South Africa in January and February 2022, respectively. Since then, they have spread to many countries and (re)increased the number of COVID-19 cases accordingly, creating a new COVID-19 wave in some countries. At present, there is limited information regarding the behavior of these subvariants, whether they have an enhanced transmissibility, immune evasiveness, or severity as compared to other SARS-CoV-2 variants. However, the rapid global dissemination of these subvariants may suggest that it could have a higher transmission rate than most, if not all, of the preexisting variants. Nevertheless, early results of recent studies have shed some light on these open questions. Yamasoba et al.31 explored the effect of several monoclonal antibodies on BA.4 and BA.5, and showed that BA.4 and BA.5 had a higher resistance to monoclonal antibody cilgavimab than BA.2. Another study by Cao et al.32 showed that BA.4 and BA.5 subvariants exerted stronger neutralization evasion than BA.2 against the plasma from 3-dose vaccination and from humoral immunity derived from post-vaccination BA.1 infections. Interestingly, they showed a contrasting result that cilgavimab could effectively neutralize BA.4 and BA.5. Next, using a deep-learning method, Chen et al.33 studied the variants infectivity and revealed that BA.4 and BA.5 were 36% more infectious than BA.2.
BA.4 and BA.5 sublineages contain some specific mutations, namely L452R and F486V (Figure 4), which are located in the RBD and not present in BA.1 (the original Omicron variant).34 As compared to BA.2, both the S-proteins of BA.4 and BA.5 have similarities with BA.2, except for the presence of del69-70, L452R and F486V. The F486V mutation in their S-proteins is known to be responsible for the viral infection.34 It was also reported that D405N (which is also carried by BA.2 subvariant) and BA.4/BA.5-specific L452R and F486V facilitated the neutralizing antibody evasion.
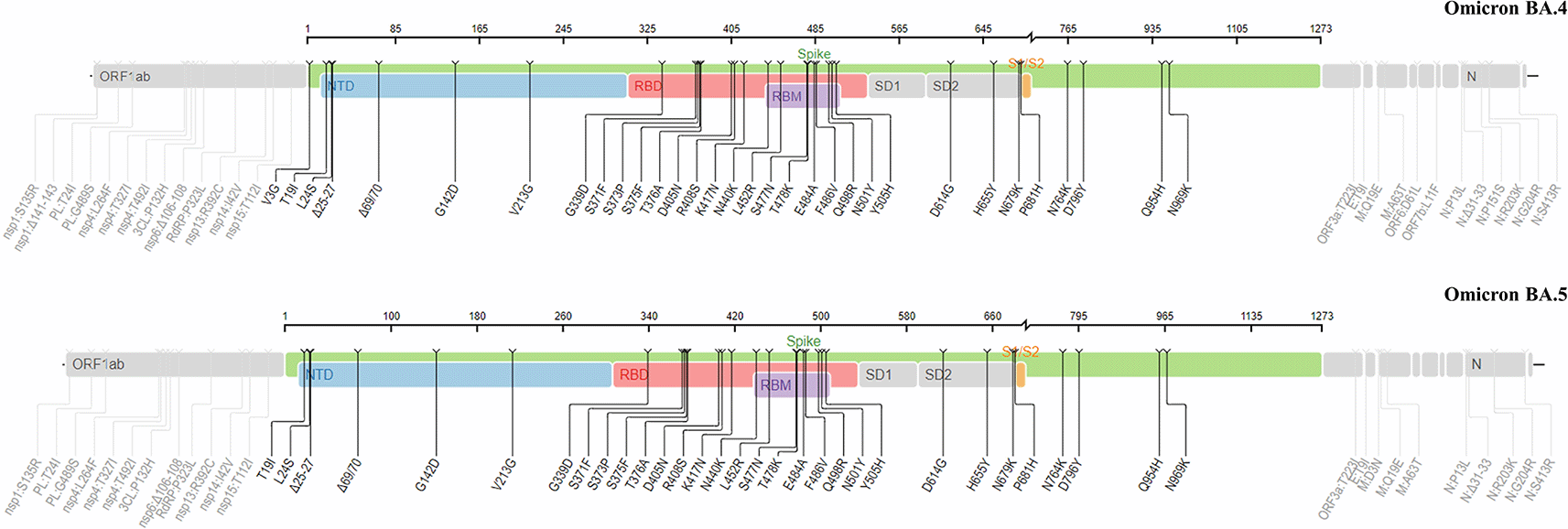
This figure was obtained from the Stanford University Coronavirus Antiviral and Resistance Database under license CC BY-SA 4.0.
The change of viral tropism in the Omicron variant is also important to scrutinize. A recent UK-based study showed that the Omicron variant was strongly associated with symptoms from upper respiratory tract more than the lower respiratory tract.35 This could be explained by the presence of two Omicron-specific mutations: N764K and N856K. Those mutations were shown to produce cleavage sites for subtilisin-kexin isozyme-1/site-1 protease (SKI-1/S1P) serine protease predominantly situated in the upper airway. Such cleavage sites are important to cleave viral envelope glycoproteins, which modulate SARS-CoV-2 replication and pathogenesis.36 Indeed, the Omicron variant was shown to replicate faster in the bronchus than in the lung parenchyma.37 Another study also highlighted that despite the similar viral replication in human nasal epithelial cultures, the Omicron variant demonstrated lower replication in lower respiratory and pulmonary cells than the Delta variant.38 Additionally, Omicron spike protein has a lower S1/S2 cleavage efficiency than the Delta variant and Omicron tends to avoid cells expressing a high level of transmembrane protease, serine 2 (TMPRSS2). This was due to Omicron’s failure in exploiting the TMPRSS2 that promotes cell entry via plasma membrane fusion. As a consequence, the cell entry of the Omicron variant was largely mediated through the endocytic pathway.38,39 The fact that SKI-1/S1P was also present in pulmonary macrophages36 could suggest the potential consequences of N764K and N856K mutations on the host innate immunity. Nevertheless, more studies are needed to confirm this notion.
Real-time reverse transcription polymerase chain reaction (rtRT-PCR) or quantitative RT-PCR (qRT-PCR) is a gold-standard diagnostic tool for identifying SARS-CoV-2, the causative agent of COVID-19. This method usually detects ≥ 2 genes of SARS-CoV-2, including ORF1ab/RdRp, N and E. 40 At present, due to the increasing incidence of Omicron variant infection, S gene with S gene target failure (SGTF) is frequently used as an indicator for screening of the Omicron variant. Most reported Omicron variant sequences include a deletion in the S gene, which can cause an SGTF in some PCR assays.
Cycle threshold (Ct) is the thermal cycle number at which the amplified DNA that shows as a fluorescent signal exceeds and thus passes the threshold for positivity. A higher Ct value means that the tool requires more copy numbers to reach the positivity threshold, indicating a lower viral concentration in the specimens. Conversely, the lower the Ct level the greater the amount of identified target ribonucleic acid (RNA) in the sample. A recent study assessing the real-time (RT) qPCR data from 10,324 specimens and comparing 97 Omicron against 107 Delta variant infections reported that the Ct values was higher for Omicron infections than for Delta infections (Omicron: Ct 23.3 vs. Delta: Ct 20.5), suggesting a lower peak viral RNA of the Omicron variant. Moreover, the clearance phase for Omicron was shorter while the clearance rate was similar with the Delta variant.41 Similarly, studies conducted in France also reported a significantly higher Ct value for Omicron infection than the non-Omicron variants.42,43 These findings suggest that the high transmissibility of the Omicron variant might not be due to a high viral load in the upper respiratory tract, despite the presence of D614G mutation which was known to increase viral replication in the upper respiratory tissue.23 Indeed, a study comparing 2,001 Alpha (median Ct = 22.0), 792 Delta (median Ct = 19.7), and 1,935 Omicron variant (median Ct = 20.8) samples reported that the Omicron infection did not have higher viral loads than those with Delta when stratified by the major PCR platforms used and by symptomatic vs. asymptomatic status. Additionally, consistent with prior studies, the study displayed higher viral loads in symptomatic patients, as compared to the asymptomatic ones. Overall, the study suggested that the rapid dissemination of the Omicron variant was not attributed to higher nasal viral loads as compared to prior variants.44 Thus, at present, the exact cause of a higher transmissibility in Omicron variant infection than other variants, including Delta, remains to be elucidated.
A recent observational study conducted in Texas, US, from late December 2021 to early January 2022 reported that the Omicron variant displayed some different clinical patterns than its predecessors.45 Compared to patients infected by Alpha and Delta variants, these patients were younger and predominantly female. The number of patients requiring hospitalization was significantly lower in Omicron than in Alpha and Delta variant infections (19.8% vs. 54.6% and 43.1%, respectively). Of those who were hospitalized, the average length of stay was significantly shorter in Omicron than Alpha and Delta (3.2 days vs. 5.1 days and 5.4 days, respectively). Moderate to severe cases (e.g., number of patients requiring extracorporeal membrane oxygenation [ECMO], mechanical ventilation and high-flow oxygenation) and mortality were also lower in Omicron infection. As expected, while Alpha and Delta variants affected mostly the unvaccinated individuals, the Omicron variant proportionally infected both the unvaccinated (44.1%) and vaccinated people (55.9%).45 These findings and comparable results from other studies46,47 confirm the hypothesis that the Omicron variant causes less severe disease, resulted in a lower hospitalization rate. Importantly, the fact that Omicron caused an increased mRNA vaccine (i.e., BNT162b2 and mRNA-1273) breakthrough incidence45 needs to be swiftly responded to by speeding up the vaccination booster campaign. The third vaccine (booster) dose has been shown to rescue and broaden the viral neutralization.48–51 A study comparing the mRNA vaccines effectiveness on 16,087 Omicron versus 4,261 Delta cases revealed that the second dose vaccine effectiveness against the Delta variant reduced from 89% to 80% after 240 days. Interestingly, against the Omicron variant, the second dose vaccine effectiveness was only 36% at day 7-59 and completely diminished after 180 days. However, the third dose (booster) vaccination recovered the vaccine effectiveness against the Delta and Omicron variants to 97% and 61% after day 7, respectively.51 Therefore, booster vaccination is expected to be beneficial to tackle this emerging COVID-19 variant.
Table 3 lists the reported clinical symptoms associated with Omicron variant infection.52–57 In most studies, upper respiratory symptoms (e.g., runny or stuffy nose, sneezing, cough and sore throat) dominated, while the lower respiratory complaint (e.g., shortness of breath) was identified in less than 20% of patients infected by the Omicron variant. Non-respiratory symptoms, such as fatigue, headache, muscle pain and fever, were also seen in some patients, although the number varies between studies. Importantly, COVID-19-pathognomonic symptoms, such as anosmia and ageusia, were only observed in a limited number of patients across studies (less than 25%). Overall, this observation is consistent with the abovementioned viral tropism of the Omicron variant.
| Symptoms | Brandal et al.52 N = 81 | Young et al.53 N = 87 | CDC Team54 N = 43 | Maisa et al.55 N = 277 | Menni et al.56 N = 4,990 | Hajjo et al.57 N = 500 | Total N = 5,978 |
|---|---|---|---|---|---|---|---|
| Norway | Singapore | USA | France | UK | Jordan | ||
| Sneezing | 35 (43%) | - | - | - | 3,143 (63%) | - | 3,178 (63%) |
| Runny or stuffy nose | 63 (78%) | 30 (35%) | 22 (59%) | 74 (27%) | 3,818 (77%) | 33% | ~4,172 (70%) |
| Loss of smell/anosmia | 10 (12%) | 3 (3%) | 3 (8%) | 23 (8%) | ~17% | 1% | ~892 (15%) |
| Loss of taste/ageusia | 19 (23%) | - | 25 (9%) | - | 44 (12%) | ||
| Cough | 67 (83%) | 39 (45%) | 33 (89%) | 143 (52%) | 2,486 (50%) | 47% | ~3,003 (50%) |
| Hoarseness | - | - | - | - | 2,145 (43%) | 9% | ~2,190 (40%) |
| Sore throat | 58 (72%) | 40 (46%) | - | 88 (32%) | 3,517 (71%) | 45% | ~3,928 (66%) |
| Shortness of breath | 10 (12%) | - | 6 (16%) | 31 (11%) | ~5% | - | ~297 (6%) |
| Reduced appetite | 27 (33%) | - | - | - | ~25% | - | ~1,275 (25%) |
| Nausea or vomiting | - | - | 8 (22%) | 20 (7%) | ~18% | - | ~927 (17%) |
| Abdominal pain | 5 (6%) | - | - | - | ~18% | - | ~904 (18%) |
| Diarrhea | - | 5 (6%) | 4 (11%) | 17 (6%) | ~19% | - | ~975 (18%) |
| Fatigue/lethargy | 60 (74%) | - | 24 (65%) | 158 (57%) | - | 32% | ~402 (45%) |
| Headache | 55 (68%) | - | - | 121 (44%) | 3,729 (75%) | 13% | ~3,970 (68%) |
| Muscle pain | 47 (58%) | - | - | 107 (39%) | ~30% | 29% | ~1,796 (31%) |
| Fever | 44 (54%) | 24 (28%) | 14 (38%) | 169 (61%) | ~35% | 48% | ~2,238 (37%) |
| Asymptomatic | - | 20 (23%) | 3 (7%) | - | - | 31% | ~178 (28%) |
Overall, the symptoms of the Omicron variant infection were different than those caused by other COVID-19 VoCs infection. For example, the ZOE COVID study in the United Kingdom56 reported that in the Delta variant infection, the loss of smell or anosmia was more prevalent (52.7% vs. 16.7%) and sore throat was less common (60.8% vs. 70.5%) than in the Omicron infection. Additionally, the duration of sickness was shorter in the Omicron infection, as well as the rate of hospitalization.56 Whilst, the Alpha variant that was phylogenetically connected with the Omicron variant8 shared many similar symptoms, including fatigue, headache, runny nose, sneezing, cough and sore throat. However, anosmia and dysosmia that were common in the Alpha variant were not frequent in the Omicron variant infection.58 Another study investigating the clinical characteristics of the Gamma variant infection in 313 healthcare workers in Brazil showed that common cold, headache, cough and sore throat were the most common reported symptoms of the Gamma infection, while anosmia and ageusia were only reported in less than 25% of the sampled individuals.59 This clinical presentation is very similar with the clinical characteristics presented by the Omicron variant, possibly due to their close phylogenetics connections.9,10
It is also crucial to acknowledge that although studies have shown compelling evidence of a less severe disease and a lower hospitalization due to Omicron infection, the high transmissibility of this variant might still impact the healthcare system readiness and increase the absolute number of hospitalization.60 These conditions would cause a severe delay on the non-COVID-19 patient care and might result in bigger indirect consequences than we expected. Also, the effect of the Omicron variant in immunocompromised individuals, and patients with concomitant diseases and comorbidities (e.g., diabetes mellitus) is still unknown. In general, these individuals have an impaired immune response against infections, a condition that favors Omicron and other COVID-19 variants. Nonetheless, the effect of Omicron on this special population is yet to be explored.
Overall, the genomic profile of the Omicron variant hinted a high affinity to ACE2 receptor, a high transmissibility and a likelihood to evade neutralizing antibodies,23 either from the previous vaccination48,61 or prior infection(s).62 This evidence suggests the need for Omicron-specific vaccines, which could provide a better protection against the disease than the currently available COVID-19 vaccines. Indeed, an Omicron-based vaccine is currently being studied in a clinical trial to evaluate the safety, tolerability and immunogenicity in healthy adults. In addition, the fact that the Omicron variant had a lower predisposition to damage lower respiratory tract and pulmonary tissue could indicate its potentially lower severity than its predecessors (e.g., Delta variant). Indeed, the less severe feature of Omicron has been seen in several observational studies. Nonetheless, the consequences of this variant on special populations (e.g., individuals with altered immune response, diabetes mellitus or advanced age) remain to be elucidated.
Regardless, implementing an effective PHSCM could still be beneficial to limit person-to-person transmission of Omicron variant. The application of well-fitting mask, imposing strict physical distancing, cough etiquette and hand hygiene, as well as avoiding crowds remain vital. The WHO has already advised to enhance surveillance with rapid testing, cluster investigations, contact tracing and quarantine, as well as case isolation to cut the chain of transmission. Only by a collaborative effort, the COVID-19 pandemic could be brought to an end.
An earlier version of this article can be found on https://www.preprints.org/ (doi: 10.20944/preprints202202.0224.v1).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the topic of the review discussed comprehensively in the context of the current literature?
Partly
Are all factual statements correct and adequately supported by citations?
Yes
Is the review written in accessible language?
Yes
Are the conclusions drawn appropriate in the context of the current research literature?
Partly
References
1. Vieillard-Baron A, Flicoteaux R, Salmona M, Chariot A, et al.: Omicron Variant in the Critical Care Units of the Paris Metropolitan Area: The Reality Research Group. American Journal of Respiratory and Critical Care Medicine. 2022; 206 (3): 349-363 Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Medical Intensivist with an area of expertise in pulmonary viral infections an ARDS
Is the topic of the review discussed comprehensively in the context of the current literature?
Partly
Are all factual statements correct and adequately supported by citations?
Partly
Is the review written in accessible language?
Yes
Are the conclusions drawn appropriate in the context of the current research literature?
Partly
References
1. Arora S, Grover V, Saluja P, Algarni YA, et al.: Literature Review of Omicron: A Grim Reality Amidst COVID-19.Microorganisms. 2022; 10 (2). PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Virology, Molecular Diagnostics, SARS-CoV-2
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 28 Jul 22 |
read | |
|
Version 1 23 Mar 22 |
read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)