Keywords
COVID-19, ACE2, Candida albicans, Aggregatibacter actinomycetemcomitans, Fusobacterium nucleatum, Veillonella parvula
This article is included in the Pathogens gateway.
This article is included in the Emerging Diseases and Outbreaks gateway.
This article is included in the Coronavirus (COVID-19) collection.
COVID-19, ACE2, Candida albicans, Aggregatibacter actinomycetemcomitans, Fusobacterium nucleatum, Veillonella parvula
We modify the manuscript according to the reviewer's suggestion in Introduction, Figure 1 legend, discussion section and the limitation of the project.
See the authors' detailed response to the review by Juni Handajani
See the authors' detailed response to the review by Jukun Song
The oral microbiota may be involved in the pathogenesis of SARS-CoV-2 infection, the causative agent of COVID-19. In addition to other oral bacteria, Candida spp., particularly C. albicans, is a keystone commensal in the human oral cavity1 that may be involved in dysbiotic events. Indeed, C. albicans is commonly reported for its relationships with known constituents of the oral biofilm in individuals with and without oral disease.2–4 As dysbiosis of the oral microbiome has been associated with inflammatory conditions in the oral habitat of COVID-19 patients with comorbidity, as reported by Bachtiar et al. (preprint),5 we assumed that fungal-bacterial interactions might also favor the establishment of SARS-CoV-2 infection. Therefore, our objective was to investigate the level of C. albicans, its pathogenicity, and to evaluate its antagonistic relationship with Aggregatibacter actinomycetemcomitans, which is a keystone pathogen associated with periodontitis in adolescents6 in the saliva of COVID-19 patients with and without diabetes. We included Fusobacterium nucleatum and Veillonella parvula as their relationship has been previously reported.7
The study was conducted at the Universitas Indonesia Hospital (RSUI), Depok, Indonesia. The eligible patients were recruited consecutively (up to 23), from August 2021 to September 2021. According to medical records, the patients had mild to moderate symptoms with clinically and laboratory confirmed COVID-19 infections at RSUI. Six subjects who visited the RSUI periodontal clinic served as a control. The average age of the participants was 45.1 ± 15.37 years old, and 10 patients had diabetes.
According to guidance provided by the Ethics Committee, written and oral information was given, after which written informed consent was obtained from all participants before enrolment in this study. The study protocol was approved by the ethics committee of Universitas Indonesia Hospital (protocol number: 2021/04/052). The protocol conformed to the criteria of the Declaration of Helsinki and the good clinical practical guidelines of the International Council on Harmonization, and this study was carried out in accordance with the guidelines provided by the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement.
Unstimulated saliva (2 ml) was collected by spitting into a sterile Falcon tube. Tongue samples were taken by swabbing the middle third of the tongue dorsum with a sterile cotton swab for a few minutes.8 The obtained samples were then put into a microcentrifuge tube. All collected samples (saliva and tongue swab) were delivered promptly to the laboratory for further processing.
Fungal genome extraction was performed using GENEzol™ reagents, (phenol, guanidine isothiocyanate solution) (Geneaid Biotech Ltd, New Taipei City, Taiwan), accordance with the protocol provided by the company. The concentration and quality of the obtained DNA were determined using Qubit assay reagents (Thermo Fisher Scientific, Waltham, MA, United States). To amplify DNA in saliva samples, we used quantitative PCR (qPCR) with specific primers for C. albicans as follows: Forward: 5′-CACGACGGAGTTTCACAAGA-3′ and Reverse: CGATGGAAGTTTGAGGCAAT-3′.9 Further, the fungus amount was calculated by plotting the cycle threshold (Ct) value against the log of a standard curve shown in Figure 1A. The standard curve was constructed using a 10-fold serial dilution of DNA extracted from C. albicans (ATCC 10231).8 The amplicon melting curves was set at 95°C for 15 seconds, 60°C for 60 secods, and 95°C for 15 seconds.
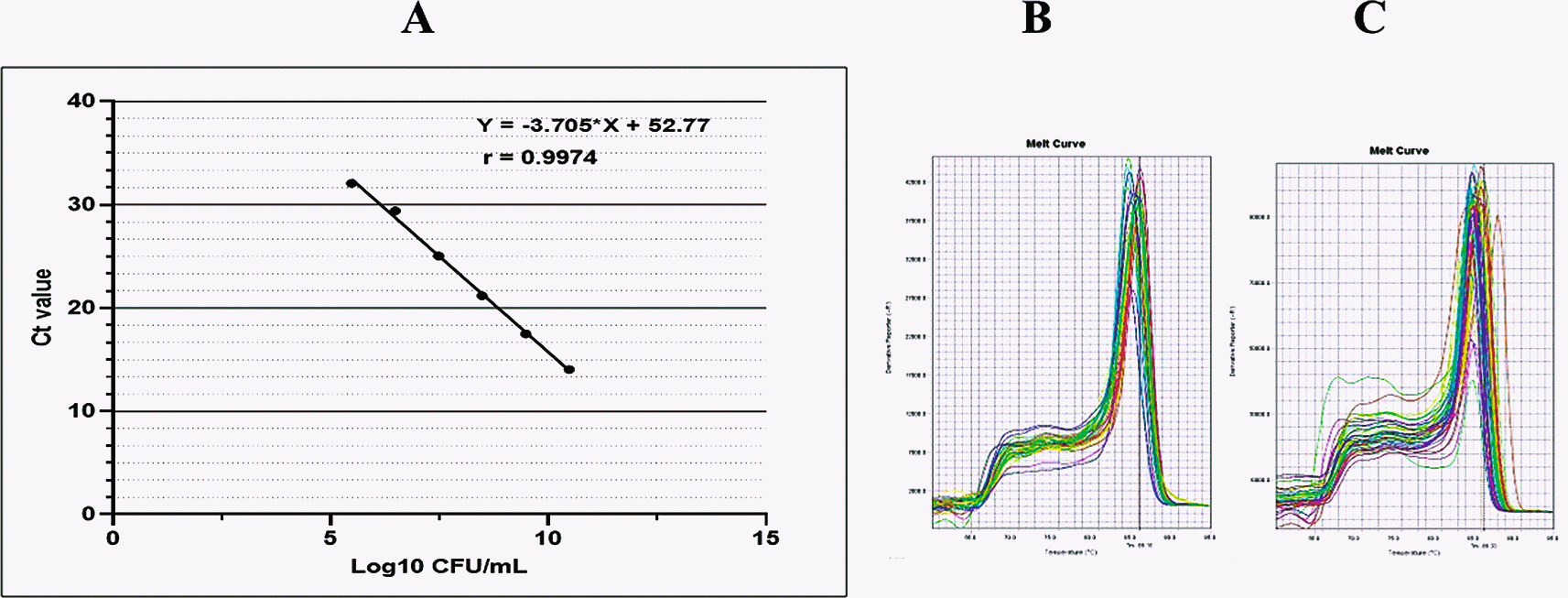
The standard curve was linear as indicated by a linear correlation (r2) of 0.9974 between the cycle threshold (Ct) value and template concentration, and a slope of -3.705.
The PCR reactions were performed at a 10-minute initial denaturation at 95°C, followed by 40 cycles of denaturation at 95°C for 15-seconds annealing at 60°C for 60 second and elongation at 95°C for 15 seconds. As shown in Figure 1B and C, the qPCR products were visualized as a melting curve, that was set at 95°C for 15 seconds, 60°C for 60 seconds, and 95°C for 15 seconds.
Bacterial DNA was extracted from the saliva samples with a similar procedure described above. The levels of A. actinomycetemcomitans, F. nucleatum, and V. parvula colonization and the total amount of bacteria in saliva were determined using qPCR with the specific primers,5 except for A. actinomycetemcomitans, for which we used oligonucleotides as reported elsewhere,10 as follows: GTGGGGAGCAAACAGGATTAG (forward) and CCTAAGGCACAAACCCATCTC (reverse).
For both C. albicans and bacterial abundance, the PCR cycling process was performed in a total volume of 10 μl (comprising 5 μl of SYBR1 Selected Master Mix (Thermo Fisher Scientific, Waltham, MA, USA), DNA template (2 μl), and primer pair solution (1 μl, 300 nM/reaction). The abundance of each bacterium was determined by using the 2-ΔΔCt method.11 ΔCt was the difference between the Ct value using the primers for each bacterium and the Ct value obtained by using the primers for total bacteria in saliva. ΔΔCt was the difference between the ΔCt of the patient and control subjects, where the value of 2-ΔΔCt shows the changes in bacterial proportion in the sample of patients relative to those of the control subjects.
For extracting total RNA, we used GENEzolTM reagent (Geneaid; Biotech Ltd, New Taipei City, Taiwan), followed by a reverse transcription kit (High-Capacity cDNA Reverse Transcription Kit, Applied BiosystemsTM). We followed all instructions provided by the kits. The resulting cDNA was amplified by qPCR with specific primers, as follows: YWP1; F: 5′-GCTACTGCTACTGGTGCTA-3′, R: 5′-AACGGTGGTTTCTTGAC-3′, HWP1; F: 5′-GCTCCTGCTCCTGAAATGAC-3′, R: 5′-CTGGAGCAATTGGTGAGGTT-3′, and ALS1; F: 5′-CAACTTGGGTTATTGAAACAAAAACA-3′, R: 5′-AGAAACAGAAACCCAAGAACAACC-3′.9
Quantitative PCR analysis was performed in triplicate on an ABI StepOnePlus Real-Time PCR System with SYBR Green PCR Master Mix (Applied Biosystems). The qPCR cycling conditions consisted of a 10-minute initial denaturation at 95°C followed by 40 PCR cycles of 15 seconds at 95°C and 1 minute at 60°C. The formula of fold change 2-ΔΔCt was used to calculate the relative mRNA expression, which was compared with that of the housekeeping gene, ACT1 with primers: F: 5′-TTTCATCTTCTGTATCAGAGGAACTTATTT-3′, R: 5′-ATGGGATGAATCATCAAACAAGAG -3′.12 The formula of fold change 2-∆∆Ct was used to calculate the relative mRNA expression of genes (ALS3, HWP1, and YWP1), which was normalized to that of the housekeeping gene ACT1.13 All values obtained from the tested patient groups were standardized and compared to the values obtained from the control subjects.
In this study, we compared the amount of C. albicans, relative abundance of bacteria, and mRNA transcription levels of the targeted genes in two groups: patients with COVID-19 with diabetes (DG) and patients without diabetes (NDG). Statistical analyses were conducted using GraphPad Prism 9.0 (GraphPad Software, San Diego, CA, USA) (RRID:SCR_002798) (An open-access alternative is the R Stats Package 4.3.0). One way ANOVA and an unpaired Student’s t-test were used to determine the p-values between and within groups, respectively. Data are presented as the mean ± standard error (SE), and p < 0.05 was considered significant. Spearman’s correlation coefficient (r) with two-tailed p-values was used to measure the degree of association between two variables tested. The line of best fit (95% confidence interval) was shown by using linear regression. The receiver operating characteristic (ROC) method was also used to determine the sensitivity and specificity of the relationship between C. albicans and A. actinomycetemcomitans as predictors of oral dysbiosis in patients with COVID-19.
As shown in Figure 2A. ACE2 mRNA expression was found in all saliva samples collected from either subject tested (DG, NDG, and control). This finding was confirmed by comparing ACE2 transcription levels on tongue surface sample (TS), where ACE2 transcription level is highly expressed in this niche.14 We observed, that mean transcription level of ACE2 detected in saliva was lower than on the TS (p < 0.05). When comparing the two group, we found that in both saliva and TS, the transcription levels of ACE2 was higher in NDG than in DG, but the difference was not significant (p > 0.05). We further determined that C. albicans was also present in the saliva and TS samples collected from all subjects. In general, the count of C. albicans in saliva samples was higher than that of TS samples (p < 0.05). Additionally, the amount (log DNA copies) of C. albicans in the saliva of DG was significantly higher than that in NDG (p < 0.05). In contrast, the different number of C. albicans on tongue surface found in either group was not significant (p > 0.05) (Figure 2B).
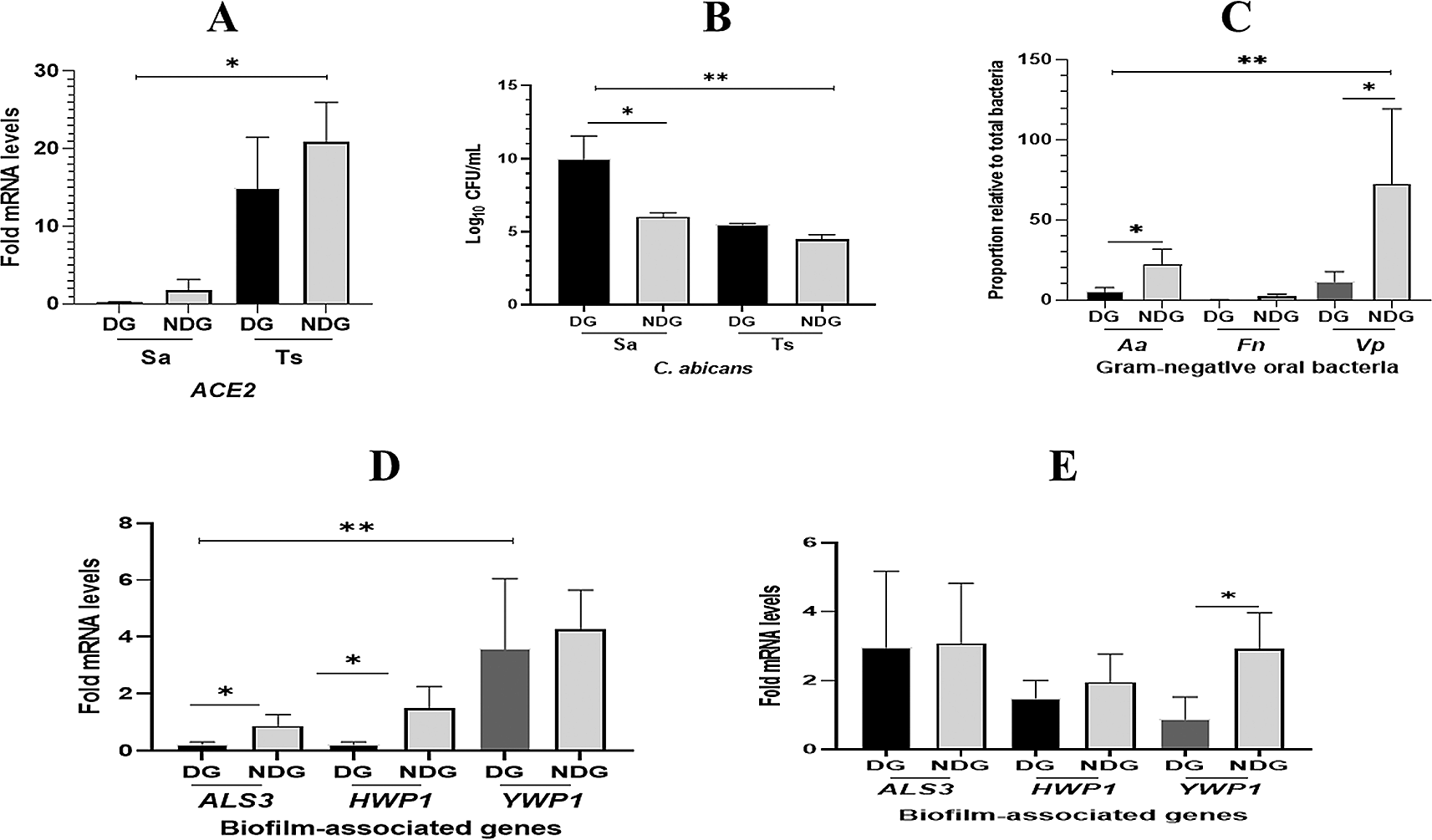
In general, the transcription level of ACE2 mRNA is higher in tongue surface (Ts) than those in saliva (Sa). Between groups, the different level of ACE2 expression is not significant (A). The number of C. albicans are significantly higher (p < 0.05) in saliva than in tongue surface samples, and C. albicans abundance is significantly higher in DG than in NDG (B). The proportion of Vp is found to be the highest compared to the other two species (Aa and Fn). Between groups, the abundance of Aa and Vp is significantly higher in DG than in NDG (C). Fold changes in gene expression detected in saliva (D) and tongue surface (E) were each compared between and within patient groups (DG and NDG). All quantitative polymerase chain reaction values were normalized according to the expression of housekeeping gene ACT1. All data are expressed as mean ± SE. *p < 0.05, **p < 0.001.
The qPCR results showed that the proportion of each bacterium in all subjects tested was lower in DG than in NDG. We found that the proportion of A. actinomycetemcomitans and V. parvula in NDG was >20% higher than that in DG (<10%). Subsequently, in either group tested, the abundance of F. nucleatum was found to be the lowest (<5%) compared to the proportion of the other two species (Figure 2C).
The qPCR results showed that the transcription levels of ALS3 and HWP1 in saliva were significantly lower than those in YWP1 (p < 0.05). We found that the transcription of both hypha-associated genes (ALS3 and HWP1) was significantly higher in NDG than in DG (p < 0.05), whereas no difference was found in the expression of YWP1 mRNA (Figure 2D). Furthermore, we analyzed the relative expression levels of each gene on the tongue surface (TS). As expected, in both groups tested, the hypha-related genes (ALS3 and HWP1) showed upregulation at a similar level. Conversely, a higher level of YWP1 mRNA expression was significantly detected on the TS of NDG subjects than in DG subjects (Figure 2E).
Moreover, as shown in Figures 3A and B, a strong negative linear correlation was observed between the abundance of C. albicans and the relative proportion of A. actinomycetemcomitans in DG (r = -0.79, p = 0.02), whereas in NDG, a low negative, non-significant correlation was observed between the two oral microorganisms (r = 0.08, p = 0.79). We noted, that in DG there was a positive but not significant correlation between the proportion of C. albicans and F. nucleatum (r = 0.05; p = 0.9) as well as with V. parvula (r = 0.24; p = 0.29) Conversely, a negative non-significant correlation was observed between C albicans and F. nucleatum/V. parvula. The correlation coefficients were r = -0.5, p = 0.08, and r = -0.41, p = 0.15, respectively (Figure 3C–F).
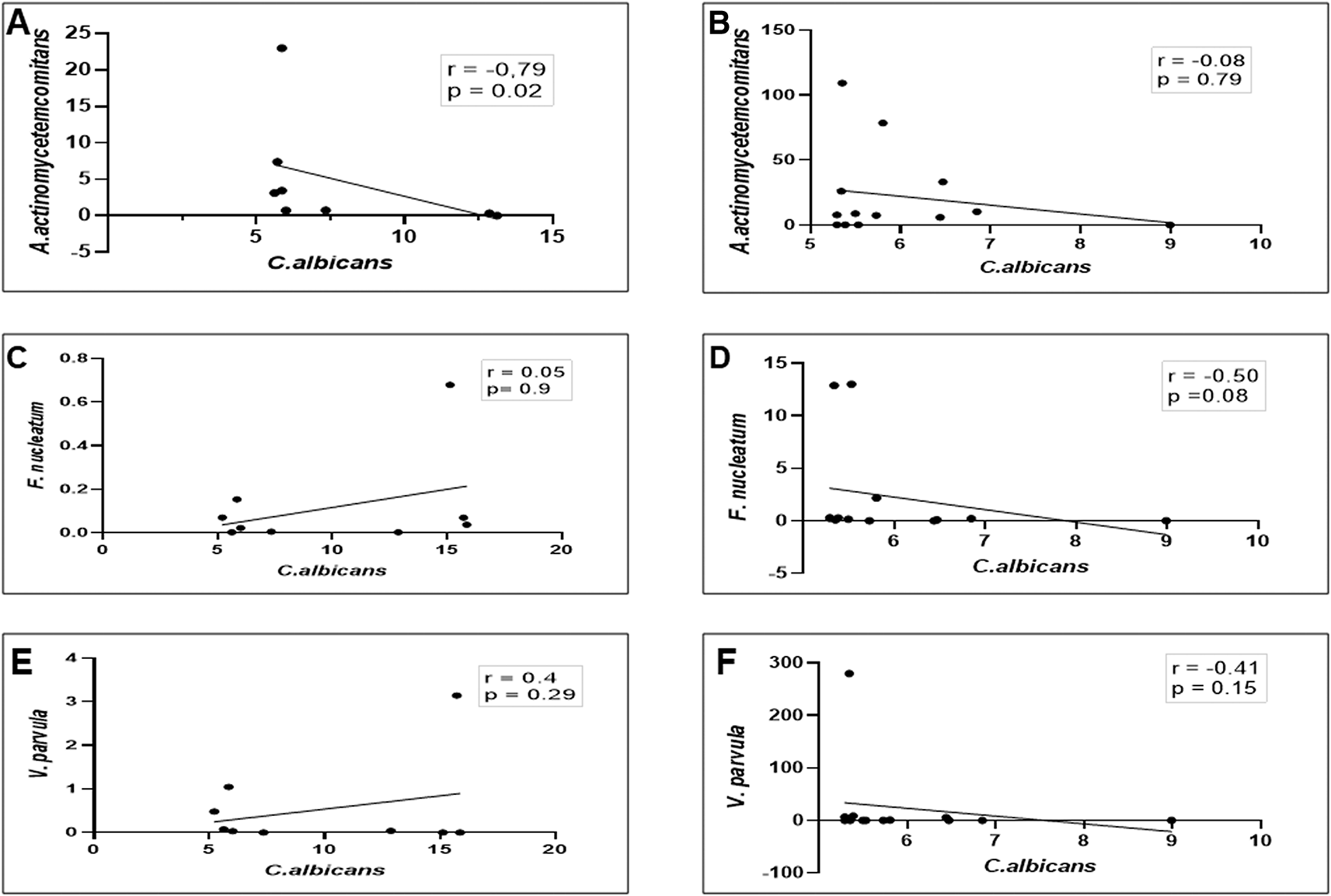
In either patient group (DG or NDG), a strong negative correlation is consistently seen between Ca and Aa (A and B), while the other two bacteria show a weak positive in DG (C and E) and negative correlations in NDG, respectively (D and F). Spearman correlation coefficient (r2) and exact p-values are given.
As SARS-CoV-2 has been consistently detected in the saliva of infected patients,15 we further examined the relationship between ACE2 expression and the relative abundance of C. albicans and A. actinomycetemcomitans in the saliva of patients with COVID-19. As shown in Figure 4A–D, a strong and significant positive correlation between ACE2 mRNA transcription and the abundance of C. albicans/proportion of A. actinomycetemcomitans was observed in DG (r = 0.81, p = 0. 01 and r = 0.75, p = 0.03, respectively). In NDG, the correlation was positive, but not statistically significant (r = 0.53, p = 0.06 and r = 0.06, p = 0.83, respectively). Based on these results, we evaluated the accuracy of the combination of ACE2 and C. albicans/A. actinomycetemcomitans relationship analyses. We revealed that the area under the curve (AUC) of the ACE2/C. albicans association, in DG was 1 (95% CI: 1 to 1, p < 0.0002; Figure 5A), and in NDG, the AUC was 0.76 (95% CI: 0.54 to 0.99, p < 0.019; Figure 5B). For the relationship between ACE2 and A. actinomycetemcomitans, in DG the AUC was 0.80 (95% CI: 0.56 to 1, p < 0.02; Figure 5C). In NDG, the AUC was 0.82 (95% CI: 0.66 to 0.98, p < 0.005; Figure 5D).
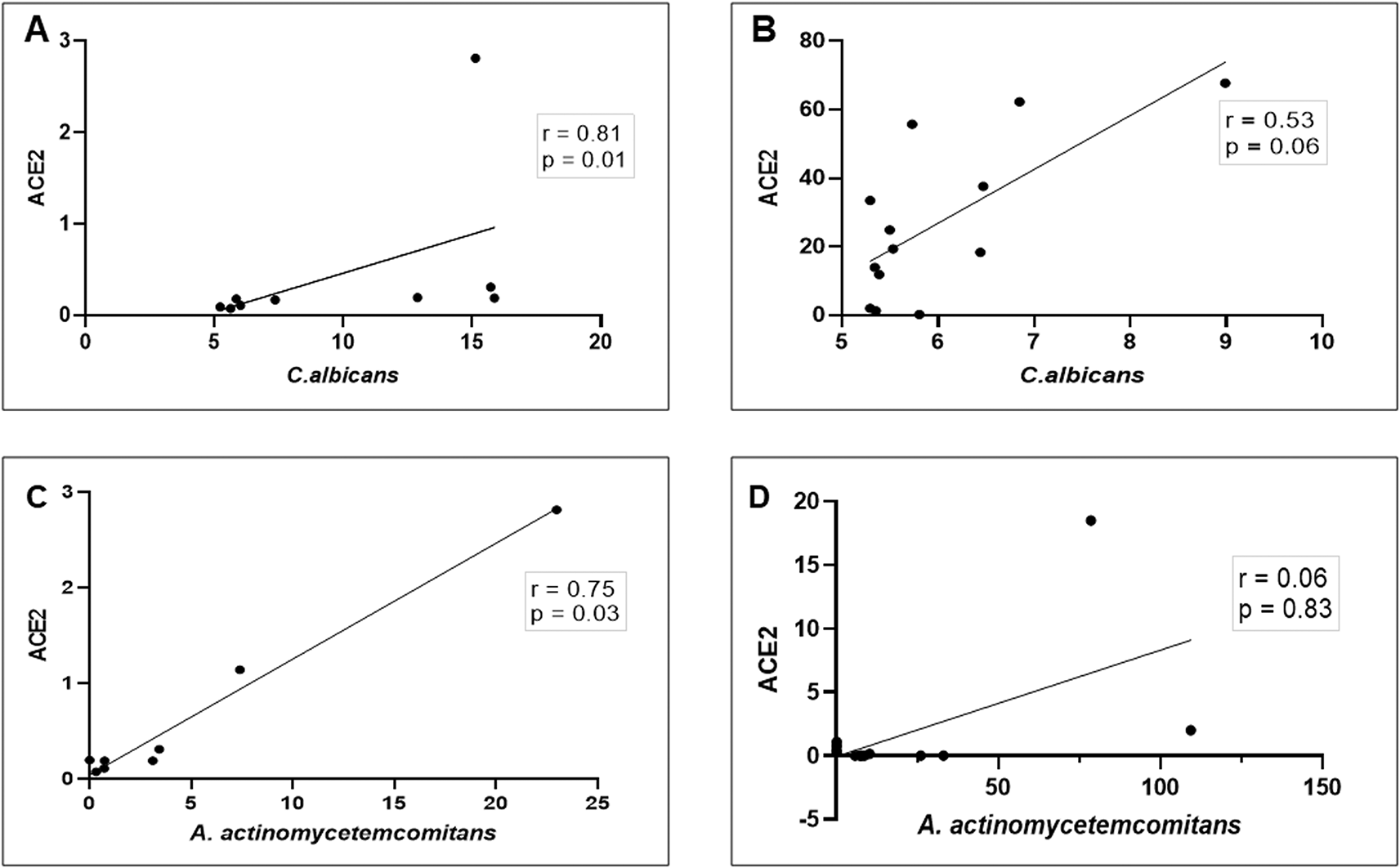
These observations indicate that in DG, the correlation between ACE2 mRNA expression and the abundance of C. albicans/A. actinomycetemcomitans is strongly positive (A and C), while in NDG the correlation is weakly positive (B and D). Spearman correlation coefficient (r2) and exact p-values are given.
This study found that ACE2 expression was detected at a lower level in saliva than on the tongue surface, indicating that saliva from patients with COVID-19 might harbor epithelial cells containing SARS-CoV-2. However, the epithelium of the tongue is likely the primary target of SARS-COV-2 in the oral cavity, which is consistent with previous findings.16,17 Therefore, although the oral environment is not the main target of SARS-CoV-2, the oral cavity could constitute both a portal of entry and a reservoir for the virus. This finding also indicated that ACE2 may be crucial for the progression and prognosis of COVID-19. Hence, exploring ACE2 expression under different physiological conditions may help predict the susceptibility of SARS-CoV-2 in different cohorts, such as COVID-19 patients with and without comorbid diabetes. The comorbidity observed in our subjects appeared to accelerate the expression of ACE2 mRNA in oral niches. Since the susceptibility to SARS-CoV-2-driven infection correlates with ACE2 expression,18 it is possible that in our subjects, either the tongue or epithelia-containing saliva have been exposed to SARS-CoV-2 infection. We assumed that this result could be linked to the clinical status of our COVID-19 patients, which, according to the patient’s medical history, only developed mild to moderate illness.19
In this study, we aimed to compare the interactions observed between C. albicans and gram-negative oral bacteria. As an important constituent of oral commensal flora, C. albicans shows a diverse inter-kingdom relationship under certain conditions, ranging from synergistic to antagonistic.20–22 The results of this study showed that the number of C. albicans in the saliva of both patient groups was increased compared to the fungal count on the tongue surface, but a significant increase was only observed in DG. This finding indicates that although saliva provides antimicrobial activity against the opportunistic oral fungal pathogen,22 this critical function is less effective in patients with COVID-19 with diabetes. Therefore, it is possible that the protective functions normally observed in saliva, such as cleansing, lubrication, and antibacterial activity, had changed in our patients with COVID-19.
Additionally, studies have shown that most patients with COVID-19 have one or more systemic (e.g., use of broad-spectrum antibiotics, use of corticosteroids, immunosuppression) or local (e.g., use of dental prostheses, reduced salivary flow due to use of medication) risk factors that favor Candida proliferation.23–25
Additionally, most participants had poor oral hygiene (not shown), and we did not measure the local risk factors. However, the high count of salivary C. albicans observed may indicate that low salivary flow rate influences salivary colonization by this fungus.26
According to the medical records, all patients had received a non-invasive supplemental device. However, we did not have any data regarding how long the patients had received the device at the time the samples were collected.
Previous studies have shown that Candida airway colonization is associated with prolonged use of mechanical ventilation and length of hospital stay.26,27 According to the medical records, all patients had received a noninvasive supplemental device. However, we did not have any data regarding how long the patients had received the device at the time the samples were collected. Additionally, although the local risk factors had not been measured, the high count of salivary C. albicans found in this study may suggest that low salivary flow rate facilitates salivary colonization by this fungus,28 as reported previously in elderly populations.8 Thus, we assumed that the physiological effects of COVID-19 on salivary gland secretion29,30 might affect the salivary flow rate of patients with COVID-19.
Additionally, we analyzed the pathogenicity of C. albicans in saliva and on the tongue surface by comparing the expression of selected biofilm-associated genes (ALS3, HWP1, and YWP1), since these genes alters the morphology of the fungus, from the yeast to hyphal form.31
Our data showed that in both patient groups (DG and NDG), the transcription levels of ALS3 and HWP1 were downregulated in saliva but conversely upregulated in tongue sample swabs. This finding suggests that the tongue is an ideal biotic surface in oral habitats for C. albicans attachment and growth. This study also revealed no difference in the pathogenicity of C. albicans (evaluated by qPCR) between DG and NDG patients with COVID-19. Hence, the results showed that the tongue surface is a better oral sample for detecting C. albicans pathogenicity in patients with COVID-19, irrespective of the diabetic condition.
ALS3 (adhesion-related gene) and HWP1 (hypha-specific gene)32 are produced predominantly during biofilm formation, while YWP1 is a yeast-associated gene involved in the anti-adhesive activity of C. albicans.31 Therefore, the higher expression of ALS3 and HWP1 mRNAs on the tongue surface indicate that this is an important reservoir of C. albicans colonization. From here the fungus may dislodge into the saliva, where the expression of YWP1 was found to be higher.
An additional phenomenon revealed in this study was the relative proportion of gram-negative bacteria in patients with COVID-19. Our data indicated that the selected oral bacteria were found in the saliva of all participants recruited in the current investigation (DG, NDG, and control). This finding suggests that the bacteria exist as normal microflora in the oral cavity, and they may have been involved in disease processes observed in our patients with COVID-19.5 The current study highlighted that in the presence of C. albicans, the most abundant gram-negative bacteria in both diseased groups (DG and NDG) were V. parvula, followed by A. actinomycetemcomitans, while F. nucleatum was the least abundant. Moreover, all species can be detected in periodontitis patients with diabetes,33 but only F. nucleatum and V. parvula have been reported to be positively associated with COVID-19-associated events.34,35 Therefore, in this study, it was deemed pertinent to assess the relationship between the proportions of these three gram-negative bacteria and the abundance of C. albicans in the COVID-19 related oral environment. Analysis of saliva samples showed that in NDG (patients without diabetes), the proportion of all selected gram-negative bacteria had a significant negative correlation with the increasing load of C. albicans DNA. Interestingly, in DG (patients with diabetes), only the proportion of A. actinomyces was consistently and significantly negatively correlated with a higher count of C. albicans. This finding supports our previous work regarding the ability of A. actinomycetemcomitans to reduce biofilm formation by C. albicans when grown in mono36 or dual species with Streptococcus mutans.6 The current findings provide additional information in vivo, whereby the salivary component in patients with COVID-19 modulates the relationship pattern shown by C. albicans when interacting with A. actinomycetemcomitans. In the present study, a negative association between the fungus and periodontal pathogen was observed by counting both microflora in saliva samples collected from patients with COVID-19. The relationship pattern may be relevant, since it not only translates closer to the real inter-kingdom relationships in vivo (oral cavity), but also demonstrates the relationships between C. albicans and other selected gram-negative bacteria in the salivary environment. From the results here, the negative association between C. albicans and A. actinomycetemcomitans observed in salivary microbiota could be multifactorial and not be simply caused by the effect of SARS-CoV-2 in the oral cavity. However, this may explain why the presence of SARS-CoV-2 in the oral cavity favors the emergence and persistence of dysbiosis in another oral niche, including the periodontal microenvironment.5 Our observations provide additional information, in which there is a strong negative correlation between the “key stone commensal” oral microflora, C. albicans1 and periodontopathogen, A. actinomycetemcomitans.37 Interestingly, we found that both C. albicans and A. actinomycetemcomitans showed a positive correlation with ACE2 expression, and a strong correlation was observed in patients with COVID-19 accompanied by diabetes. Therefore, it was deemed relevant to assess these relationships by determining the sensitivity and specificity of the association. Using ROC curve analyses, we found that the antagonistic relationship between C. albicans and A. actinomycetemcomitans and its respective correlation with ACE2 expression had an obvious effect in distinguishing between DG and NDG and could be used as a biomarker with a certain degree of accuracy. Thus, the correlation of ACE2 expression with C. albicans/A. actinomycetemcomitans had a higher predictive value for oral dysbiosis in patients with COVID-19 with diabetes.
The literature shows that C. albicans is implicated in oral diseases, including caries,2 periodontitis,38 denture stomatitis,39 and endodontic lesions.40 We suggest that there is a synergistic relationship between the receptor for SARS-CoV-2 entry (ACE2) and C. albicans/A. actinomycetemcomitans as the underlying mechanism of oral dysbiosis in COVID-19 patients with diabetes. These relationships may be crucial to the persistence of C. albicans and A. actinomycetemcomitans as part of the oral commensal flora and may potentially contribute to the progression of polymicrobial infection-associated dysbiosis under COVID-19 conditions.
This study has limitations. It was not possible to include all potential confounding variables. Unlike the abundance of C. albicans, we used the relative abundance of each targeted bacterium species as a proportion11 rather than the actual levels. Moreover, due to the sample size is too small, the statistical power might lack of power.
This study revealed that the saliva of patients with COVID-19 with diabetes retained a special relationship between SARS-CoV-2 host entry and the oral dysbiotic atmosphere represented by a unique pattern of C. albicans and A. actinomycetemcomitans. This relationship could be associated with the existence of SARS-CoV-2, but it is necessary to consider the complicity of diabetes.
Open Science Framework: Underlying data for ‘ACE2 expression in saliva of patients with COVID-19 and its association with Candida albicans and Aggregatibacter actinomycetemcomitans’. https://doi.org/10.17605/OSF.IO/ENFY319
This project contains the following underlying data:
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
Written informed consent for publication of the patients’ details was obtained from the patients.
All the authors would like to acknowledge all the study participants, Director of RSUI and the Ethical Clearance Committee for providing permission and ethical clearance to conduct this research project. We acknowledge the contribution provided by NR. Utami, A. Deviana, AD. Andiryani, for technical lab., and gathering data.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: I am committed to the mechanism research of oral and maxillofacial tumors.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Oral Biology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 12 Sep 22 |
||
|
Version 1 23 May 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)