Keywords
lung cancer, EGFR, KRAS, TP53, survival rate.
This article is included in the Oncology gateway.
lung cancer, EGFR, KRAS, TP53, survival rate.
In the latest version of this article, there are some additional points regarding the possibility of mutations in several different genes, especially for EGFR and TP53 simultaneously. A more in-depth discussion on the prognostication of lung cancer patients with concomitant gene mutations based on existing studies in other countries is also added. Even so, there were no concurrent mutations in this gene in the sample of this research study. In the discussion section, this research again clarified that mutations in certain genes and exons can determine a patient's prognosis that is worse than others. Regarding the management of lung cancer patients included in this study, each patient has a personalized combination of treatment and is discussed further in the discussion section. Meanwhile, regarding the prognostication study of lung cancer patients with standardized management based on one particular treatment in the Indonesian population such as TKI (tyrosine kinase inhibitor) will be included in further research.
See the authors' detailed response to the review by Darren Wan-Teck Lim
Recent molecular studies have focused on the genetic alterations and epigenetic impacts in the pathogenesis of lung cancer.1 Several tumor suppressor genes and oncogenes have been identified and altered the micro and macro environment related to lung carcinogenesis.2 This was a multistep process where the oncogenes mutation initiates and triggers conversions of the normal epithelium to cancer cells due to environmental factors, particularly cigarette smoke.1 P53 and KRAS genes play crucial roles in maintaining cells integrity that affects the early stage of lung carcinogenesis, particularly in lung cancer-related tobacco exposure.3 Recent studies showed the mutations of KRAS and TP53 mutations have been related to shorter overall survival and progression-free survival rates in lung cancer patients, whether in early or advanced stages.4,5 In contrast, EGFR mutations, as one of the most common oncogenic mutations together with targeted therapy management, gives longer survival times than for subjects with wild-type mutations who received systemic chemotherapy.6–8
However, there has been a surge of EGFR resistance around the world. The concurring mutations with other oncogenes and tumor suppressor genes have been identified as the cause of EGFR resistance.3–5,9,10 Unfortunately, there is still limited data regarding oncogenes and tumor suppressor gene mutations in Indonesia. This study aimed to assess the role of oncogenes and tumor suppressor genes in determining the survival rate of lung cancer patients in the population of North Sumatra.
This was a retrospective cohort study conducted in the Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, Universitas Sumatra Utara during the period of 2017–2020. All subjects were enrolled in the study via several cancer-referral hospitals in North Sumatra in collaboration with the Faculty of Medicine, Universitas Sumatra Utara. The criteria for inclusion in this study were subjects aged 18 or over and diagnosed with lung cancer confirmed by histopathology preparations from bronchoscopy, open biopsy, thoracotomy, and trans-thoracal lung biopsy. The number of cells in one section of the paraffin block must be a minimum of 50 cells. All medical records were then observed to assess the demographic and clinical characteristics and followed the progressivity of disease based on RECIST criteria.11 Subjects’ deaths were recorded in their medical records in the case of subjects who passed away in hospital, while others were assessed based on phone calls from the subjects’ relatives addressed in medical records. The exclusion criteria of this study were an inadequate amount of cancer cells in a paraffin block, incomplete medical records, and difficulties in assessing the subjects’ relatives.
All the procedures of this study were approved by the Ethics Committee of the Faculty of Medicine, Universitas Sumatra Utara with reference number 124/KEP/USU/2020. Written informed consent was obtained from all the subjects or their relatives. All the pathology specimens received permission for genetic analysis from the Pathology Department at each collection site.
DNA was extracted from Formalin-Fixed Paraffin-Embedded using Quick-DNA FFPE mini prep (Zymo Research, USA) and sliced using microtome in 1–2 slices and 4 μm thickness. After being deparaffinized, digested, and purified, the DNA was eluted by a DNA elution buffer. The PCR was performed using MyTaq HS Red Mix (Bioline, UK) with the following process: denaturation, annealing, and elongation. DNA was then amplified using certain primers (Table 1) and run in 2% agarose electrophoresis in Tris Buffer EDTA pH 8.0. Sequencing analysis was carried out via two different methods. EGFR and KRAS were converted to reverse complement and analysed using ClustalW in BioEdit Sequence Alignment Editor 7.2.5. The reverse sequence was also compared to human reference genome NG_007524.2 (LRG_433) using BLASTn NCBI. The BLASTn results were also aligned with corresponding Coding Sequences (CDS). Lastly, the sequence was analysed using FinchTv 1.4 Software using reverse complement views. Meanwhile, TP53 genes were analysed using Unipro Ugene v. 38.1 software (RRID: SCR_005579) and compared with standard reference NG-017013.2. Samples in which there was a change in protein base analysed using Nucleotide Basic Local Alignment Search Tool (BLASTn) were then compared with several TP53-specific databases including SNP (dbSNP), Cosmic (assessing somatic mutation), ClinVar (Clinical variations, assessing the correlation of mutations with clinical profile).
As shown in Figure 1, a total of 52 subjects completed the procedures of this study with most of the subjects being male (75%), aged over 60 years (53.8%), heavy smokers (75%), with adenocarcinoma type lung cancer (69.2%) stage IVA (71.25%). Further characteristics were as described in Table 2. From sequencing analysis results no KRAS exon 2 mutations were detected, while EGFR mutations were detected in 6 subjects, consisting of 5 subjects who had exon 19 mutations, 1 subject with intron 19 mutations, and 2 subjects with exon 21 mutations. TP53 mutations were seen in 5 subjects, of which 2 subjects had missense mutations of exon 5, 1 subject showed silent mutations, 1 subject with missense exon 8mutations, and 1 subject had nonsense exon 8 mutations. Associated with coexisting genetic mutation simultaneously, such as EGFR and TP53 mutations in an individual, were not found in this study.
Survival analysis for EGFR mutations is depicted in Figure 2. EGFR mutations showed a significant increase in both overall survival (OS) and progression-free survival (PFS) with p=0.001 for OS and p=0.018 for PFS (Log-Rank Test). Five of six subjects showed mutations in exon 19 and 2 subjects showed double mutations in exon 19 and exon 21. Further analysis also showed significant improvements in OS and PFS of both single and double mutations of EGFR while double mutations showed higher OS and PFS compared with single mutations. Detailed comparisons of EGFR mutations are described in Table 3.
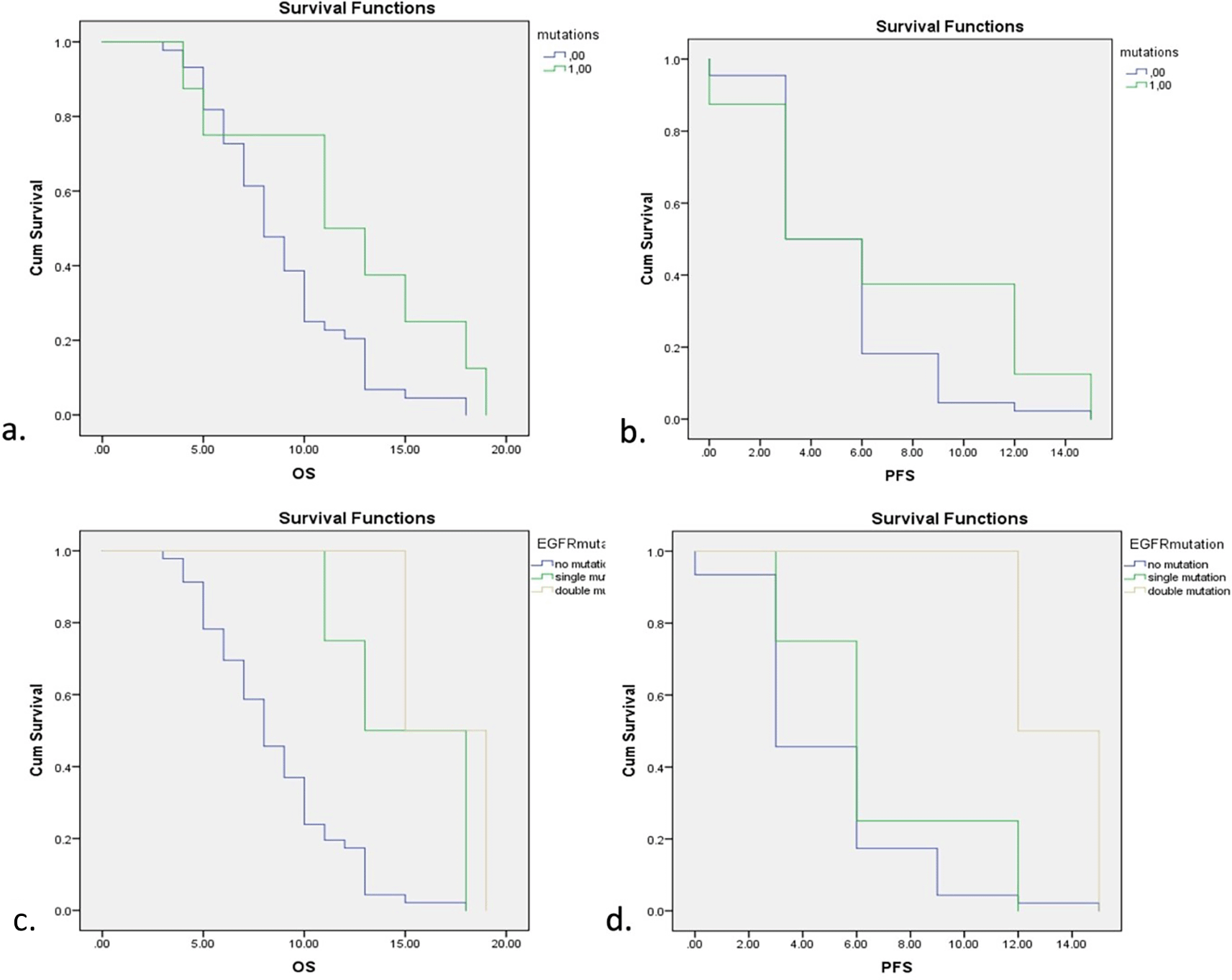
a. Overall Survival (OS) with and without EGFR mutations. b. Progression-free Survival (PFS) with and without EGFR mutations. c. OS with different types of EGFR mutations. d. PFS with different types of EGFR mutations.
| Mutations | Overall survival | p-value | Progression-free survival | p-value |
|---|---|---|---|---|
| EGFR mutations | 15 | 0.001* | 6 | 0.018* |
| Exon 19 | 15 | 6 | ||
| Intron 19 | 18 | 12 | ||
| Exon 21 | 15 | 12 | ||
| Double mutations | 15 | 12 | ||
| TP53 mutations | 7 | 0.148 | 3 | 0.070 |
| Exon 5 | 9 | 3 | ||
| Exon 6 | 5 | 3 | ||
| Exon 8 | 7 | 6 | ||
| Missense | 7 | 3 | ||
| Silent | 5 | 3 | ||
| Nonsense | 9 | 6 | ||
| No mutations | 8 | 0.037* | 3 | 0.196 |
Survival analysis was presented in OS and PFS curve in Figure 3. Generally, subjects with TP53 mutations had a lower survival rate including OS and PFS compared with non-mutations. Subjects with TP53 mutations had 7 months of OS, 2 months less than subjects without TP53 mutations (p=0.148). This study also compared median survival rates for different types of TP53 mutation including non-mutations, missense mutations, nonsense mutations, and silent mutations as follows; 9 months, 7 months, 9 months, and 5 months. Nevertheless, the association between median survival rates and different types of TP53 mutation was not significant in statistical analysis with a p-value of 0.245. Patients with TP53 mutations also had a three months’ shorter PFS compared to those with non-mutations (3 months vs 6 months; p=0.07). After further analysis according to different types of mutation, the PFS of subjects with missense mutations and silent mutations was 3 months, while for subjects with non-mutations and nonsense mutations it was 6 months. These different mutations were also insignificant in relation to median PFS with a p-value of 0.107.
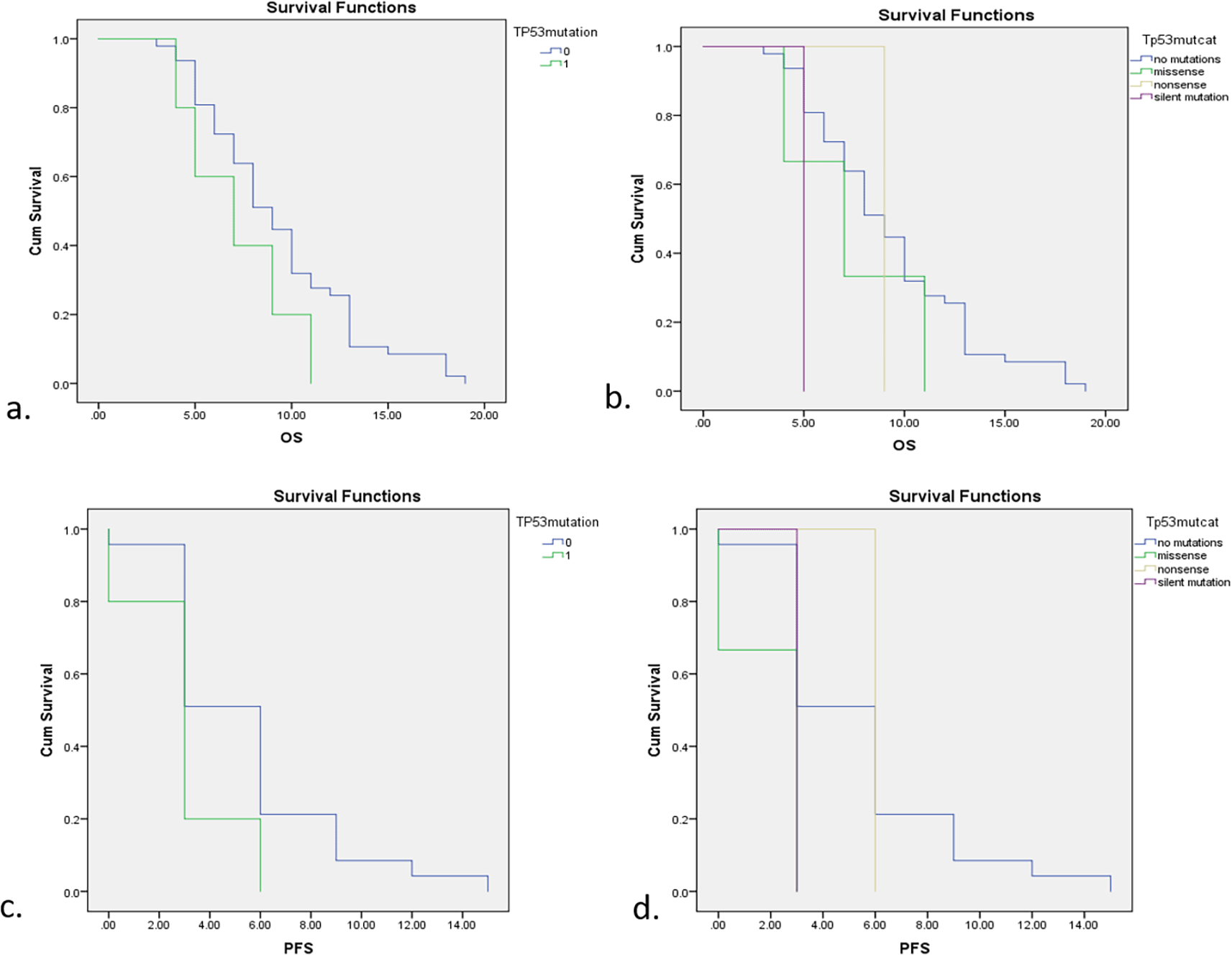
a. OS between subjects with and without TP53 mutations. b. OS with different types of TP53 mutation. c. PFS between subjects with and without TP53 mutations. d. PFS with different types of TP53 mutation.
The survival rates assessed in this study were overall survival (OS) and progression-free survival (PFS) and Table 3 shows the comparisons of OS and PFS in subjects with EGFR, TP53, and no mutations. EGFR mutations showed a significant increase in both OS and PFS compared with no mutations, while TP53 was a poor predictor of lung cancer patients with shorter survival rates.
KRAS mutation is one of the common oncogenes mutations that are related to poor prognosis of lung cancer and is strongly associated with smoking.12–19 This study follows on from a study by Soeroso et al. which analyzed KRAS mutations in the population of North Sumatra.20 However, there were no KRAS exon 2 mutations among the subjects of this study. The scanty population of KRAS mutations in Asia is not completely understood, although recent data has shown that Asia has a lower prevalence of KRAS mutations compared with worldwide data.21,22 In Asian populations, KRAS mutations were present in 4.3%–10.5% of the population, while data shows that KRAS mutations were present in 30–40% of populations worldwide.23 In addition, Syahruddin et al., as the first study to identify KRAS mutations in lung cancer in Indonesia, showed a lower incidence of KRAS mutations compared with populations worldwide (7% vs 31%).24,25 The occurrence of KRAS mutations also often coincides with mutations in other oncogenes or tumor suppressor genes. Co-mutations between KRAS and TP53 were the most prevalent and were related to the poorest outcomes in all pharmacological approaches including targeted therapy, chemotherapy, and radiotherapy.25 However, studies by Fang et al. and Dong et al. showed a better outcome in administering immune checkpoint inhibitors in a patient with KRAS and TP53 mutations compared with KRAS mutations alone or with no mutations at all.26,27 On the other hand, KRAS mutations were among the most common factors related to EGFR-TKI resistance. Several studies reported the concurring EGFR-KRAS mutations in 15–20% populations28–31 while KRAS mutations were presumed to be independent factors in resistance to EGFR-TKI treatment. The absence of KRAS mutations in this study directs the pulmonologists to analyze other factors which might contribute to resistance to EGFR-TKI treatment in the population of North Sumatra and other populations with a low prevalence of KRAS mutations, despite the limitations of facilities in detecting KRAS mutations in certain populations.
KRAS mutations can be detected in cytology and histopathology examinations following the RT-PCR or DNA Sequencing method.32 The type of KRAS mutation will be identified using the DNA-Sequencing method where a change in GGT to TTG is addressed as G12C,33 the most common mutation of KRAS related to longer survival rate compared with other mutations.25 The second most common KRAS mutation type is in codon 12 changes in GGT to GTT, referred to as G12V mutations.33 In this study, we examined the histopathology preparations from the paraffin block for all subjects diagnosed with lung cancer, and from the PCR test it was found that almost all subjects showed expressions of RAS protein in exon 2 codon 12-13. Nevertheless, there was no change in protein base with DNA sequences showing GGT-GGC known as normal RAS protein expressions. This finding showed that the Sanger sequencing method carried out in this study is still reliable in identifying the KRAS mutations, although the variations in North Sumatra populations showed no mutations.
Role of the EGFR mutations in lung cancer has been described in previous studies.8,34,35 EGFR mutations treated with EGFR-TKI-targeted therapy showed a significantly improved survival rate with lung adenocarcinoma.7,35,36 However, the multiple mutations are not completely understood. The various studies tried to identify the impact of double mutations or triple mutations of EGFR in clinicopathology characteristics in lung cancer patients.37,38 All previous studies showed poorer outcomes with multiple mutations compared with single EGFR mutations.37,38 Nevertheless, in this study, double mutations of exon 19 and exon 21 showed a better outcome in both OS and PFS compared with single mutations of exon 19. Unfortunately, this study was a small size study, so it is still difficult to draw general conclusions concerning the wider population. In the previous studies, the double mutations consisted of exon 20 T790M, which has been identified as primary factors in 1st and 2nd EGFR TKI-resistance.38 Barnet et al. also showed shorter PFS in a patient with co-mutations of exon 19 with noncommon EGFR mutations including exon 20, T90M, and PIK3CA.39 This differs from the current study, which looked at exon 19 and exon 21 mutations. A larger scale of genetic studies is needed to evaluate the impact of double mutations in the treatment of lung adenocarcinoma, both in receiving targeted therapy and systemic chemotherapy.
In addition to the oncogenes mutations mentioned above, mutations of TP53 as one of the most common tumor suppressor gene mutations associated with resistance to cancer therapy.40,41 Having a role as the guardian of the genome, the TP53 gene maintains the integrity of the genome by modifying, stabilizing, and preventing cell proliferation in response to cellular stress.42–45 This study found five subjects with TP53 mutations and showed a shorter survival rate in both OS and PFS. When different kinds of mutation were analyzed in depth, missense and silent mutations showed shorter OS and PFS compared with nonsense mutations. However, the small number of subjects analyzed in this study might bias the effect due to individual variations. On a larger scale of different types of TP53, a missense mutation is the most common TP53 mutation,46 particularly in tobacco-related lung cancer.46,47 According to the latest evidence, missense mutation has a “gain-of-functions” effect, leading to an increase in the expression of cancer cells.48,49 In contrast, Halvorsen found that missense mutations showed higher OS compared with other types of TP53 mutations although, in general, TP53 mutations showed a higher Hazard Ratio (HR) compared with no mutations.46
Related to the previously described genetic mutations, comutation of genes, is associated with lung cancer cases with worse prognosis. Associated with genetic mutations that are found simultaneously, such as EGFR and TP53 mutations simultaneously in an individual sample were not found in this study in North Sumatra. Seeing most of the studies in various parts of the world related to this shows that TP53 mutation is an important marker of poor prognosis and predictor of lung cancer, especially in cases of NSCLC with EGFR mutations.50–54 By comparing TP53 mutation group with TP53 wild-type group, a recent meta-analysis on this issue confirmed worse OS of TP53 in EGFR mutant lung cancers by 1.7 times in comparison to wild-type group (hazard ratio 1.73, 95% CI 1.22-2.44, P=0.002).54
Patients with TP53 mutation on exon 5, exon 7, exon 8 and exon 9 demonstrated better prognosis than exon 4, exon 6, mutation of unknown site and multiple mutated patients. Different EGFR mutations in patients shared different types of TP53 mutation. The “poor” TP53 mutation (exon 4, exon 6, mutation of unknown site and multiple mutation of TP53) and “better” TP53 mutation (exon 5, exon 7, exon 8 and exon 9 mutation of TP53) showed different prognostic values with almost the same trend in different EGFR mutated groups. TP53 wild type patients demonstrated the best prognosis, good mutated patients demonstrated middle prognosis and poor mutated patients demonstrated the worst prognosis in EGFR mutations.51,52,54
Regarding the management of lung cancer patients with different type of mutations in the sample of this study, each sample has received personalized treatment and indeed not all lung cancer patients receive tyrosine kinase inhibitor (TKI) drugs. The management of lung cancer patients in the sample is quite diverse covering every aspect from surgery, chemotherapy, to targeted therapy. These recent studies have indeed shown positive results in lung cancer patients, especially in adenocarcinoma patients treated with EGFR-TKI-targeted therapy, so that prognostication studies based on genetic mutations for this treatment need to be carried out, especially in Indonesia with a diverse population.7,35,36 In connection with standardized lung cancer prognostication studies based on management such as tyrosine kinase inhibitors which are also reviewed based on genetic mutations, will be carried out in future studies. This research is the first research data that adequately represents the survival rate of lung cancer patients in Indonesia which has been reviewed based on genetic mutations in North Sumatra and in general in Indonesia.
As oncogene activations occur, TP53 activates and arrests the cell cycle in both G1 and G2, allowing sufficient time for DNA repair.55,56 Along with RAS mutations, p53 is a multifunctional protein which alters cell regulations, cancer cell transformations, and vascular invasion in all solid cancers including lung cancers. Therefore, oncogenic activations and over-expressions of p53 genes are independent predictors of poor prognosis in NSCLC. In this study, there were no co-mutations of TP53 with any of EGFR or RAS mutations meaning it is difficult to evaluate the interactions of concurring mutations, whether they have mutual or opposite effects.
Other data highlighted in this study is the lower number of mutations of oncogene and tumor suppressor gene in North Sumatra. A large cohort study in Norway showed 38% of subjects had positive mutations of KRAS in non-small cell lung cancer patients, while in Chen’s study involving a lower number of subjects KRAS mutations accounted for 11% of all mutations with a higher prevalence among smokers (20–30%) compared with those who never smoked (7–13%).57 This is in contrast this to study which found no KRAS mutations in subjects diagnosed with lung cancer. EGFR was the most common oncogene mutation, particularly in Asia. The median global prevalence of EGFR in the advanced stage of NSCLC was 33.07%58 while 47% of Asians showed EGFR mutations.57 The latest data on EGFR mutations in Indonesia also revealed EGFR mutations in 44.4% of 1874 cytological specimens diagnosed with EGFR. In this study, just 11.5% of subjects were positive with EGFR mutations with allele-specific for exon 19 and 21 and DNA sequencing. Furthermore, TP53, as the common tumor suppressor gene accounted for more than 50% in NSCLC40,57,59 was only detected in 9.6% of the subjects in this study. A definite mechanism explaining the lower prevalence of both oncogenes and tumor suppressor genes may not be understood yet. Lung carcinogenesis is a complex process characterized by the after-effects of genetic variations as the individual process works synergistically with environmental exposure.60,61 Long durations of exposure to carcinogens might alter DNA methylation, resulting in the conversion of normal cells to cancer cells. Therefore, a better understanding of genetic variations and individual vulnerability of lung carcinogenesis may provide better anti-cancer research.
Lung cancer is a type of cancer with the highest prevalence in the world. Even during the current COVID-19 pandemic, cancer patients including lung cancer are a population that is quite susceptible to COVID-19 infection and lately this is something that deserves special attention and is being widely discussed. Based on the calculation of the prevalence of lung cancer in cancer patients with COVID-19, it was found that 20.23% of cancer patients with COVID-19 were lung cancer patients.62 With an in-depth study of oncogenes and tumor suppressor genes related to lung cancer, it is hoped that it can provide clinical implications, especially related to the prognosis of lung cancer patients.
Although this study has several limitations including the small samples size and the limited survival aspect, this is the first study to identify TP53 mutations in lung cancer patients in Indonesia and the first study to identify the DNA sequencing of common oncogenes and tumor suppressor genes in the population of Sumatra. Further multi-center study involving larger sample size is needed to identify the prevalence of oncogenes and tumor suppressor genes to actualized personal therapeutic approach for cancer patients, particularly in a developing country such as Indonesia.
Figshare: Underlying data for “The role of oncogenes and tumor suppressor genes in determining survival rates of lung cancer patients in the population of North Sumatra, Indonesia”, https://doi.org/10.6084/m9.figshare.19522468.63
In addition, this study adds Research Resource Identifiers (RRIDs) from these research resources including the following software tools and data:
1. Human lung carcinoma (RRID:CVCL_5791)
2. FFPE/Formalin-Fixed Paraffin-Embedded (RRID:SCR_001307)
http://www.bioconductor.org/packages/release/bioc/html/ffpe.html
3. FinchTV (RRID:SCR_005584)
4. ClustalW (RRID:SCR_017277)
5. BLASTn (RRID:SCR_001598)
6. Unipro UGENE (RRID:SCR_005579)
Data are available under the terms of the Creative Commons Universal License (CC0 1.0 Public domain dedication).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Lung cancer
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Lung cancer
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Thoracic oncology.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 15 Jun 23 |
read | |
|
Version 1 28 Jul 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)