Keywords
Fraxin, Quercetin, Anti-Cytokine Storm, RAW 264.7 Murine Macrophage Cell Line, Lipopolysaccharide, Proinflammatory Cytokines, PPAR Γ, TLR-4, Tnfα, IL1β, IL6, Synergistic Combination, MTT Assay
This article is included in the Cell & Molecular Biology gateway.
Cytokine storm syndrome (CSS) is a leading cause of morbidity and mortality in patients with late-stage coronavirus disease 2019 (COVID-19), causing multiple organ failure and death. According to prior research, fraxin, and quercetin have anti-inflammatory, antioxidant, antimicrobial, and antiviral properties. Therefore, this study aimed to investigate the anti-cytokine storm activity of fraxin and quercetin, their combination, and the molecular mechanism behind this activity in Lipopolysaccharide (LPS)-induced RAW 264.7 cells.
LPS-induced macrophage cells were treated with fraxin, quercetin, or their combinations at various doses. Cytotoxicity and cytokine release were evaluated, and gene expression analyses were performed. An enzyme-linked immunosorbent assay was used to quantify the levels of proinflammatory cytokines, interleukin 1 beta (IL1β), interleukin 6 (IL-6), and tumor necrosis factor-α (TNF-α), and RT-PCR was used to measure the gene expression of PPAR-γ and Toll-like receptor 4 (TLR-4) concerning GAPDH as a reference gene.
The results revealed a slight decrease in cell viability only when higher concentrations were applied to the cells. Fraxin, quercetin, and their combination reduced the generation of proinflammatory cytokines. The combination (fraxin + quercetin (FQ)) reduced the levels of IL-1β, IL-6, and TNF-α by 56.2%, 58.5%, and 70.6% respectively, compared to the LPS-only control; pretreatment of cells with farxin, quercetin, and their combination resulted in significant inhibition of TLR-4 gene expression by 89%, 82%, and 93% respectively, compared to the control (P˂ 0.05); FQ upregulated PPAR-γ expression up to 60-fold compared to the control, while fraxin and quercetin increased PPAR-γ by 17.6 and 8.6-folds, respectively.
Based on these findings, fraxin, quercetin, and their combination were able to mitigate cytokine release and improve the levels of gene expression involved in their pathways, making these agents and their combination candidates for further investigation in in vivo settings to expand knowledge about their kinetics and dynamics.
Fraxin, Quercetin, Anti-Cytokine Storm, RAW 264.7 Murine Macrophage Cell Line, Lipopolysaccharide, Proinflammatory Cytokines, PPAR Γ, TLR-4, Tnfα, IL1β, IL6, Synergistic Combination, MTT Assay
The revised version of this work includes the following changes:
1- Rephrasing the research title for clarity and relevance
2- Rephrasing and corrections in the language for clarity
3- The addition of a "study design flow diagram" for better understanding
4- Deletion of unnecessary parts in the methods
5- Summarizing the majority of the methodology part to the most relevant points
6- Additional future research recommendations
See the authors' detailed response to the review by Fan Jiang
See the authors' detailed response to the review by Dr. Amany Ghazy
A cytokine storm is a condition of uncontrolled systemic hyperinflammation caused by excess cytokines, leading to multiorgan failure.1 Cytokine storms may occur for many reasons, including malignancy, rheumatoid arthritis, and sepsis. Recently, cytokine storms were found to be related to mortality and morbidity in many cases of coronavirus disease 2019 (COVID-19).2 Since coronavirus disease is characterized by hyperinflammation and an excessive immune response, the need to develop anti-cytokine drugs has increased.3 Lipopolysaccharide (LPS), a component of the outer membrane of gram-negative bacteria, signals toll-like receptor 4 (TLR 4) to activate macrophages, which stimulates several intracellular signaling pathways, including those for nuclear transcription factor kappa-B (NF-B) and mitogen-activated protein kinases (MAPKs). Interleukin IL-6, IL-1, and tumor necrosis factor (TNF-α) are proinflammatory cytokines activated macrophages release.4
Fraxin, a coumarin derived from the plant Fraxinus and Cortex fraxin, is referred to as 7,8-Dihydroxy-6-methoxy coumarin, 8-D glucopyranoside.5 Fraxin possesses different pharmacological activities, including anticancer, antiviral, anti-inflammatory, and antioxidant.6 For this vast potential, fraxin is a target for further immunomodulating studies.
Quercetin is a bioflavonoid widely distributed in apples, berries, grapes, and onions. Quercetin reported in previous studies to have a wide range of biological actions, including anti-inflammatory properties due to the inhibition of inflammation-related enzymes, cyclooxygenase (COX), and lipoxygenase (LOX).7 RAW 264.7, a standard monocyte/macrophage cell line, is mainly used to study the anti-inflammatory activity of plant-derived extracts and their active constituents by evaluating the reduction in the production of inflammatory mediators, cytokines, and chemokines in LPS-stimulated RAW 264.7 cells (RAW 264.7 a macrophage cell line that was established from a tumor in a male mouse induced with the Abelson murine leukemia virus).8
Peroxisome proliferator-activated receptor γ (PPAR-γ) is a nuclear hormone receptor and a ligand-activated transcription factor family member. Increasing evidence indicates promising anti-inflammatory properties of cancer cells exerted by activating PPAR-γ with synthetic ligands.9
PPAR-γ agonists have been thought to inhibit the production of monocyte inflammatory cytokines and the expression of inducible nitric oxide synthase (iNOS), which has been observed in response to synthetic anti-diabetic thiazolidinedione drugs (such as BRL 49653 and ciglitizone), and negatively regulates the expression of proinflammatory genes and suppresses tumor cell growth.10 The anti-inflammatory effect derived from the activation of PPAR-γ is likely the result of inhibition of NF-kB, STAT signaling pathway, and MAPK signaling pathway. The inhibition of pro-inflammatory transcriptional factors and signaling pathways results in a reduction in the transcription of genes encoding pro-inflammatory cytokines (e.g., TNF-α, IL-1β, IL-6, IL-12), chemokines, and adhesion molecules. Additionally, it enhances the release of IL-10 and suppresses macrophage polarization toward the M1 phenotype (pro-inflammatory) while promoting the M2 phenotype (anti-inflammatory, tissue repair).11,12
Drug combinations have been previously used as a new approach for treating many diseases. Their beneficial effects appear to be enhancing pharmacological activity and minimizing the dose to avoid any unwanted side effects of drugs without compromising their efficacy.13 Furthermore, previous literature mentioned that both fraxin and quercetin possess some antioxidant and anti-inflammatory activity in different disease models. This research was designed to investigate the cytokine release inhibitory activity of fraxin and quercetin from the LPS-induced murine macrophage RAW 264.7 cell line, the possible mechanism underlying it, through changes in expression of TLR4 and PPAR-γ signaling pathways, and whether exhibiting a potential synergy as combination using iso-bolographic analysis, based on the median effect principle.14
This study was designed to evaluate the cytotoxicity, anti-inflammatory activity, and molecular effects of fraxin, quercetin, and their combination (FQ) in LPS-stimulated RAW 264.7 macrophages, with dexamethasone used as a positive control, as illustrated in Figure 1. The investigations followed the guidelines established by the Ethics Committee of Al-Nahrain University, College of Medicine (approval number Nah. Co. Pha.12 on 27 June 2022).
Quercetin hydrated 2-(3,4-dihydroxy phenyl)-3,5,7-trihydroxy-4Hchromenen-4-one dihydrate (purity ≥ 96%), fraxin (7,8-Dihydroxy-6-methoxy coumarin-8-beta-D-glucoside) (purity ≥ 98%), dexamethasone (purity ≥ 98%), and lipopolysaccharide (LPS) (Escherichia coli, 055: B5) were purchased from Hangzhou-Hyper Chem. Limited/China, dimethyl sulfoxide (DMSO) from Thomas Baker/India, Mouse Interleukin1β (IL-1β), (IL-6), and (TNF-α) enzyme-linked immunosorbent assay (ELISA) kits were purchased from MyBiosource, USA, RAW 264.7, (TIB-71) murine macrophage cell line (ATCC® TIB-71™), and Dulbecco’s modified Eagle’s medium (DMEM) from American Type Culture Collection (ATCC, USA), fetal bovine serum (10% FBS), Trypsin- EDTA, penicillin/streptomycin solution from Capricorn Scientific/Germany, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) kit from MyBioSource, USA; TRIazol® reagent (Invitrogen); RT-PCR primers: glyceraldehyde 3-phosphate dehydrogenase (GAPDH), PPAR-γ, and TLR-4 from OriGene/USA, LightCycler® FastStart™ SYBR® Green master kit/Roche, Germany, Revert AidTM first strand Complementary Deoxyribonucleic acid (cDNA) synthesis kit/Thermo Scientific, USA.
Cell culture
Mouse monocytes/macrophages (TIB-71™) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Capricorn Scientific, Germany) supplemented with 10% fetal bovine serum (FBS) and antibiotics (penicillin 10 IU/mL, streptomycin 10 μg/mL, amphotericin B 0.025 μg/mL) at 37 °C in a humidified atmosphere with 5% CO2.
Cell authenticity was confirmed by morphological assessment and growth curve analysis, and cultures were routinely screened for mycoplasma contamination using Hoechst staining after each passage. Cells up to passage 10 were used in all experiments. For assays, cells were seeded in six-well plates at a density of ~6 × 104 cells/mL and incubated for 2–3 days until reaching 70–80% confluence.
Cell viability in RAW264.7 macrophages was assessed using the MTT assay following treatment with quercetin, fraxin, and their combination (FQ). RAW264.7 cells were seeded at a density of 1 × 104 cells/well in 96-well plates and allowed to adhere and grow for 24 h. After this period, the medium was replaced, and cells were exposed to serial dilutions (200, 100, 50, 25, 12.5, and 6.25 μg/mL) of fraxin, quercetin, or FQ (1:1 ratio). Each concentration was tested in triplicate. Two hours post-treatment, lipopolysaccharide (LPS, 1 μg/mL) was added to each well, and the cells were incubated for an additional 24 h. Subsequently, 20 μL of MTT solution (5 mg/mL) was added to each well, followed by a 4 h incubation at 37°C in a 5% CO2 atmosphere. The medium was then discarded, and 150 μL of dimethyl sulfoxide (DMSO) was added to dissolve the insoluble formazan crystals formed by viable cells. Absorbance was measured at 540 nm using an ELISA plate reader (Human®, USA). Cell viability was calculated relative to untreated control cells, which were considered to have 100% viability.11,15 The percentage of viable cells was determined using the following formula:
Experiments were performed in triplicate, and the data are presented as mean ± standard error of the mean.
Quercetin hydrated 2-(3,4-dihydroxy phenyl)-3,5,7-trihydroxy-4Hchromenen-4-one dihydrate (purity ˃ 96%), Fraxin (7,8-Dihydroxy-6-methoxy coumarin-8-beta-D-glucoside) (purity ˃ 98%), and LPS (lipopolysaccharide) (Escherichia coli, 055: B5) were all purchased from Hangzhou-Hyper Chem. Limited/China. Both quercetin and fraxin were dissolved using DMSO (Dimethyl sulfoxide, from Thomas Baker/India), and LPS was dissolved and diluted in PBS for preparation of 1 μg/ml solution. Each agent (quercetin and fraxin) was dissolved in DMSO and then diluted to a final volume with (Mg2+, Ca2+)-free PBS buffers at pH 7.4 to prepare a stock solution of 1 mg/ml for each fraxin and quercetin. Serial dilutions were freshly prepared on the same day of the experiment from the stock solution. Agents were tested at concentrations of 200, 100, 50, 25, 12.5, and 6.25 μg/ml, and for FQ (half the concentration was tested for each agent in the same well), cells were supplemented with 200 μl of fresh medium along with the tested agent. The concentration of DMSO used (<0.1%) did not influence the performed assays.
The MTT assay results were analyzed using the isobologram method and the median-effect/combination index (CI) equation as described previously. Dose–response curves were generated for each compound individually and in combination across a range of concentrations. In this analysis, D represents the dose, and Dm corresponds to the median-effect dose (IC50). The parameters of the dose–effect relationships were calculated using the CompuSyn software, which also provided the CI values based on the general combination index equation. Interpretation of the CI values was as follows: CI < 1 indicates synergism, CI = 1 indicates an additive effect, and CI > 1 indicates antagonism.14
The inhibitory effects of fraxin, quercetin, and their combination (FQ) on cytokine release were evaluated in RAW 264.7 macrophages, with dexamethasone serving as the positive control. Proinflammatory cytokines (IL-1β, IL-6, and TNF-α) were quantified following LPS stimulation (1 μg/mL). RAW 264.7 cells were seeded in 96-well plates at a density of 1 × 104 cells/well and incubated for 24 h. Cells were then pretreated with fraxin (25 μg/mL), quercetin (12.5 μg/mL), or FQ (6.25 μg/mL), concentrations selected based on MTT assay results. Dexamethasone (5 μg/mL) was included as a reference control. All treatments were performed in triplicate, while untreated cells served as the negative control. After 2 h of pretreatment, LPS was added to each well, and cells were incubated for 24 h at 37 °C in a 5% CO2 humidified incubator.16,17 Supernatants were collected, centrifuged at 2000 × g for 10 min, and analyzed for cytokine levels using ELISA kits (MyBioSource, USA) specific for IL-1β, IL-6, and TNF-α. All reagents and dilutions were prepared according to the manufacturer’s instructions. Cytokine concentrations were determined by fitting sample absorbance values (OD measured at 450 nm) to the standard curve generated from known concentrations, and results were expressed as pg/mL.
RAW 264.7 cells were seeded into 6-well plates at a density of 1 × 106 cells/well and incubated for 24 h. Cells were then pretreated in triplicate with fraxin (25 μg/mL), quercetin (12.5 μg/mL), or their combination FQ (6.25 μg/mL). After 2 h of pretreatment, LPS (1 μg/mL) was added, and cells were further incubated for 24 h at 37 °C in a 5% CO2 humidified incubator. Following treatment, cells were washed three times with PBS, and growth medium was removed. Total RNA was extracted by lysing cells with 1 mL of TRIzol™ reagent (Invitrogen, Thermo Fisher Scientific, USA) according to the manufacturer’s protocol.18 The concentration of extracted mRNA was determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, USA). An aliquot of the measured mRNA of each sample concentration was made using nuclease-free water to make the final concentration of mRNA in all samples 0.125 μg/ml.
The two-step SYBR Green RT-qPCR method was used to measure the relative gene expression of the targeted genes (PPAR-γ and TLR4) in the RAW264.7 cell line. Quantitative PCR was performed in a final volume of 20 μL, consisting of 18 μL SYBR Green PCR master mix (containing nuclease-free water, forward and reverse primers, SYBR Green I dye, Taq DNA polymerase [1 U], 1.25 mM MgCl2, PCR buffer, and 100 μM dNTPs) and 2 μL of cDNA template. In a preliminary step, cDNA was synthesized from total RNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions.
Primers used for amplification, and their sequences (5′–3′) are listed in Table 1.
Amplification was initiated with an initial denaturation at 95 °C for 3 min, followed by 40 cycles of 95 °C for 25 s and 55 °C for 25 s. To confirm specificity, a melting curve analysis was performed from 60 to 94 °C with a ramp rate of 1 °C/s. Relative gene expression was calculated using the 2-△△Ct method, normalized to GAPDH as the reference gene, and expressed as fold-change compared to the control group.19
All tests were performed in triplicate, and the results are presented as mean ± standard error of the mean (SEM), analyzed by one-way ANOVA followed by Tukey post hoc test for multiple comparisons. SPSS (RRID: SCR_013726) version 25 was used for statistical analysis; P < 0.05 was considered significant.
The MTT assay determined the cell viability of RAW 264.7 cells; Figure 2 illustrates the effects of fraxin, quercetin, and FQ on cell viability (expressed as a percentage compared to the control-untreated cells-considered 100% cell viability) in the presence of LPS. A reduction in cell viability was noticeable with higher concentrations of both fraxin and quercetin. While FQ exhibited the highest cytotoxicity among all three treatment groups, the viability of cells was decreased in a dose-dependent manner.
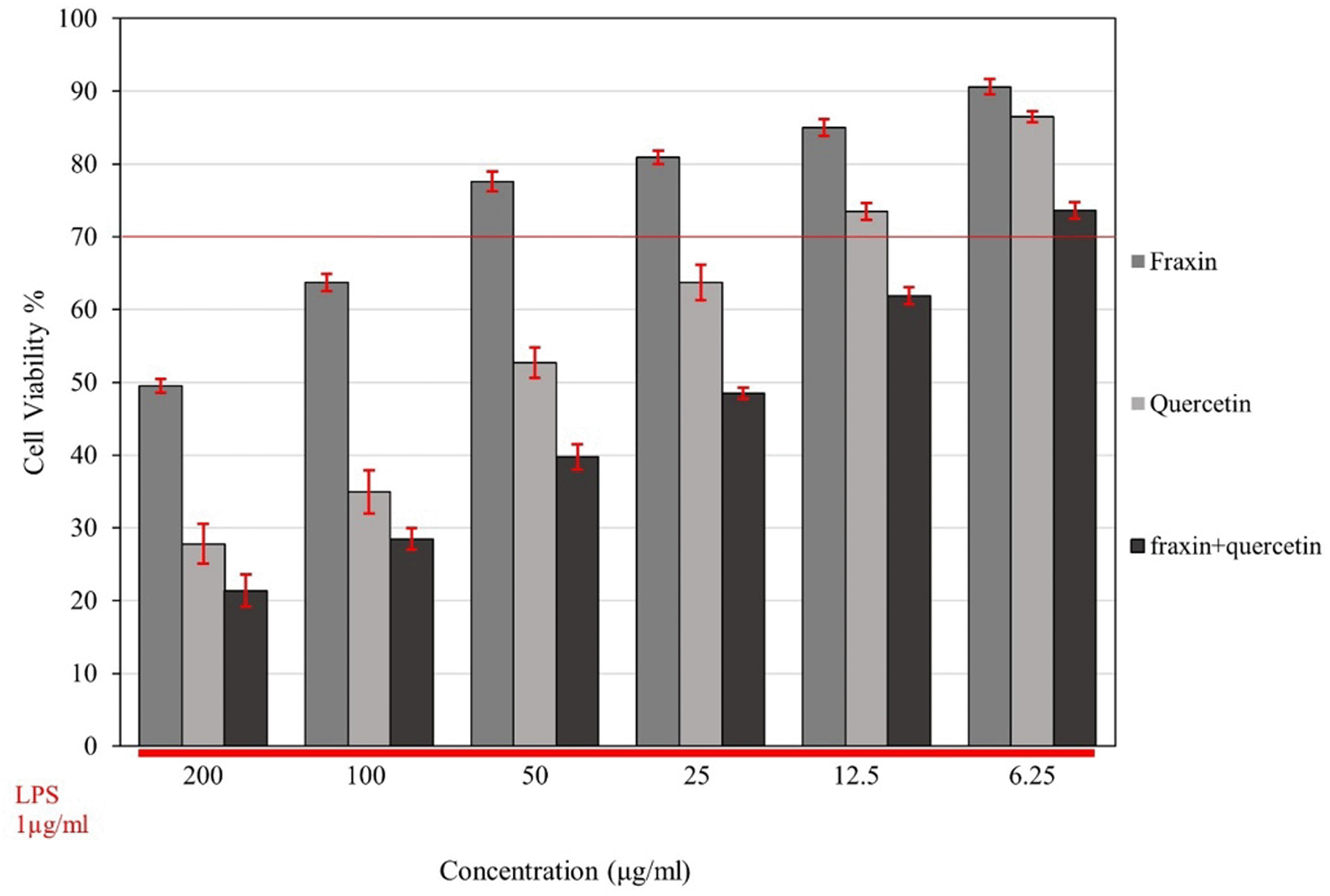
Cell viability was determined using an MTT assay. The lower red line indicates that cells were subjected to different concentrations of all treatment groups with the presence of lipopolysaccharide (LPS) (1 μg/ml), and the upper red line represents 70% cell viability in all treatments. Cell viability was expressed as a percentage compared with the control, which was considered 100% cell viability, and data are presented as mean ± SEM.
Half-maximal inhibitory concentration (IC50) values were 248, 54, and 26.5 for fraxin, quercetin, and FQ, respectively. Based on these results, the threshold for cell viability was set at 70% or higher for further evaluation of the inhibitory activity of cytokine release,8 and the concentrations (25, 12.5, 6.25 μg/ml) for fraxin, quercetin, and FQ were selected, respectively, for the evaluation.
The combination index (CI) for FQ in a 1:1 ratio was calculated using CompuSyn based on the MTT results. Dose-effect and median-effect curves were plotted for each drug and its combination. The Results showed that FQ in a 1:1 ratio exhibited synergism, with CI values of 0.297 at IC50 and CI values of 0.409, 0.333, 0.332, 0.267, 0.265, and 0.276 at the following concentrations FQ (200, 100, 50, 25, 12,5, 6.25 g/ml).
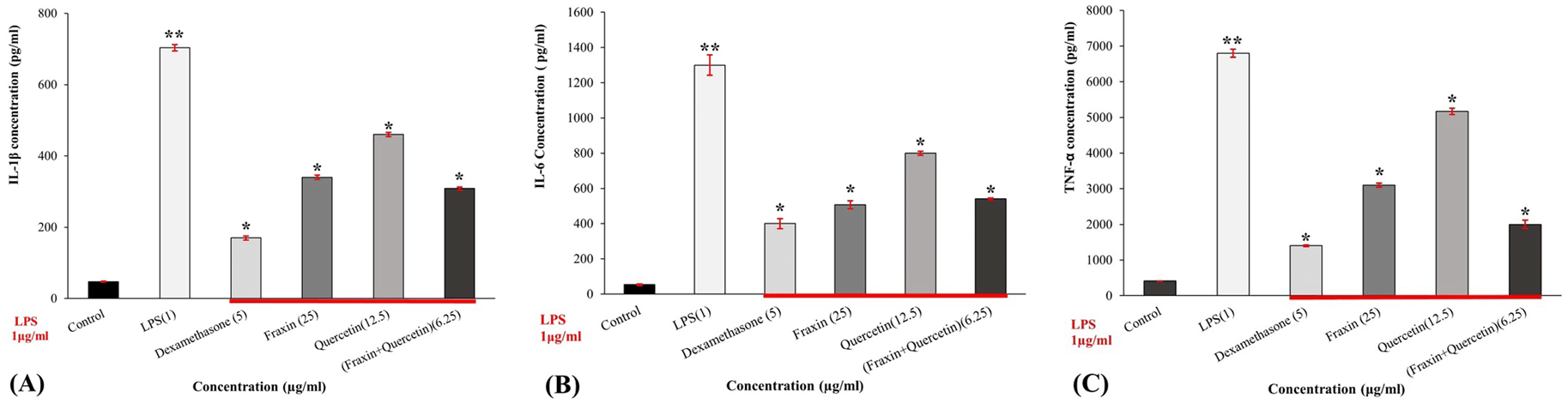
Fraxin, quercetin, and FQ in concentrations of 25, 12.5, 6.25 μg/ml, respectively, significantly suppressed the production of IL-1β, IL-6, and TNF-α (P ˂ 0.01) in a dose-dependent manner when compared to control (cells treated with LPS only). LPS significantly upregulated production of proinflammatory cytokines compared to the control group (P ˂ 0.05). The highest inhibition activity was recorded with dexamethasone (5 μg/ml) (positive, treated group) which significantly (P ˂ 0.05) suppressed IL-1β, IL-6, and TNF-α by 75.7%, 69%, and 79% respectively, compared with the LPS-treated control. In the case of the combination, FQ, the levels of IL-1β, IL-6, and TNF-α were reduced by 56.2%, 58.5%, and 70.6% respectively, suggesting it is more effective in inactivating cytokine production than each drug alone. The results are shown in Figure 3A, B, and C).
When compared to the control, pretreatment of RAW 264.7 cells with farxin (25 μg/ml), quercetin (12.5 μg/ml), and FQ (6.25 μg/ml) for 2 hours before LPS (1 μg/ml) resulted in significant (P ˂ 0.05) suppression of TLR-4 gene upregulation by (89%, 82%, and 93%, respectively). Treatment with LPS activated the TLR-4 pathway, as shown in Figure 4A, and treatment with fraxin, quercetin, and FQ successfully counteracted the stimulatory impact of LPS on RAW 264.7 cells. Furthermore, compared to either treatment alone, the combination synergistically reversed the impact of LPS on cells.
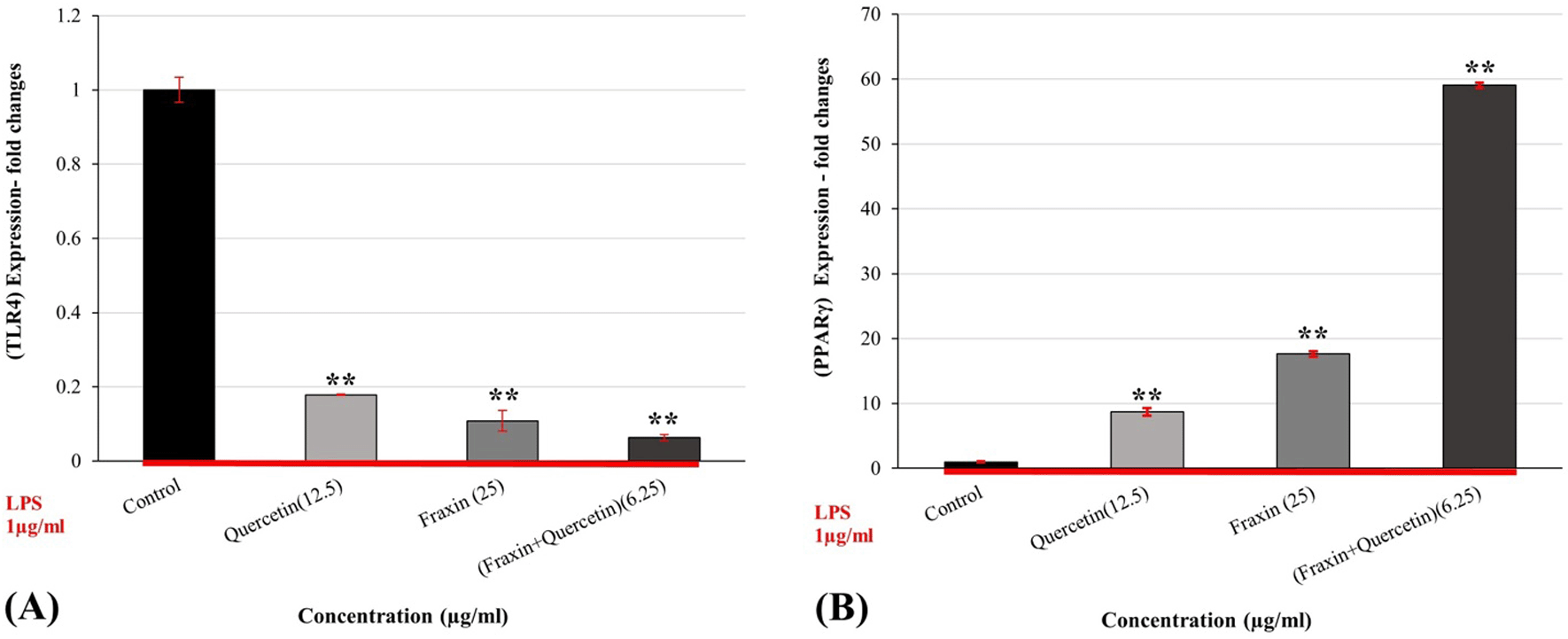
While Figure 4B reveals that there is a considerable stimulatory impact on the PPAR-γ pathway, this effect is shown as enhanced gene expression. FQ increased up to 60-fold relative to the control, whereas fraxin and quercetin (17.6, 8.6-folds, respectively) decreased proinflammatory cytokines (Figure 4B), indicating a mechanism by which fraxin, quercetin, and their combination reduce proinflammatory cytokines.
The devastating epidemic caused by SARS-CoV-2 in 2019 prompted researchers to make a considerable effort to search for a possible solution to limit infection. Following the demonstration of the pathological role of the “cytokine storm”, evidence for a cytokine release syndrome can be seen in increased proinflammatory cytokines in late-stage COVID-19. As seen in previous epidemics caused by SARS-CoV and MERS-CoV, dysregulated cytokine production and an influx of inflammatory myeloid cells can cause lung infiltration, septic shock, respiratory failure, acute respiratory distress syndrome (ARDS), multiorgan failure, and death.20,21 Gram-negative bacteria’s outer membrane lipopolysaccharide (LPS) is used in inducing a cytokine storm model both in vivo and in vitro studies. The stimulation of macrophages with LPS can cause the excessive release of proinflammatory cytokines by activating the nuclear factor κB (NF- κB) and mitogen-activated protein kinase (MAPK) signaling pathways, increasing (COX-2) and (iNOS).22 RAW 264.7 cells induced by LPS is the most widely used model for evaluating anti-cytokines in vitro. For centuries, plants have been used as a natural remedy for numerous illnesses. Hong et al. (2012) reported a dose-dependent reduction in cell spreading and pseudopodia production after treatment with an ethanol extract of Fraxinus rhynchophylla bark on LPS-stimulated macrophages.23 Whang et al. (2005) suggested in their study that fraxin and fraxin-related chemicals improved cell survival rate in human umbilical vein endothelial cells (HUVECs) when exposed to hydrogen peroxide (H2O2) mediated oxidative stress; other previous studies discussed the effect of quercetin on cell migration, which plays a vital role in the development of cancer.24
Quercetin strongly inhibited LPS-induced macrophage adhesion and migration in a dose-dependent manner.25 Previous research has highlighted the various biological activities of fraxin and quercetin, including anti-inflammatory and antioxidant effects, raising the need for additional investigation into their role in cytokine storms. Our study observed that fraxin, quercetin, and fraxin + quercetin exerted low cytotoxic activity on RAW 264.7, and only when cells were exposed to higher concentrations of fraxin and quercetin, which was in agreement with a study by Cui et al. that suggested that only the highest concentration of quercetin reduced macrophage viability when administered together with LPS (1 μg/mL).25 Other previous studies compatible with our study support that the presence or absence of LPS did not compromise the cell viability of RAW 264.7.23,26
Li et al. (2019) concluded that fraxin confers protection against LPS-induced lung injury and the inflammatory response in A549 cells.26 In an approach to modulate virus hyperinflammation such as chronic systemic symptoms, the anti-inflammatory effects of quercetin were investigated in mouse macrophage cells exposed to polyinosinic-polycytidylic acid (poly (I:C) as an experimental model for viral inflammation by Kim YJ and Park W. (2016). They found that quercetin might suppress poly (I:C)-induced inflammation by reducing the levels of inflammatory mediators.27
Our findings support previous research that found that both fraxin and quercetin were effective at suppressing the release of proinflammatory mediators from LPS-induced RAW 264.7, the mechanism underlying which may be related to interference with various inflammatory signaling pathways, including the TLR signaling system. Following their activation, proinflammatory molecules (such as IL-1, IL-6, and TNF-α) are abundantly generated, and NF-κB phosphorylation, nuclear translocation, and upregulated transcription of proinflammatory factors are all results of TLR-4 activation.22,28
In numerous macrophage models, PPAR-γ has been demonstrated to exert anti-inflammatory effects by suppressing the expression of multiple pro-inflammatory genes, including IL-6, TNF-α, and IL-12.29 From earlier studies, fraxin, isolated from the roots of Ulmus macrocarpa Hance, significantly suppressed the expression of iNOS and COX-2, increased PPAR-γ expression, activated the nuclear factor erythroid 2-related factor 2/heme oxygenase-1 (HO-1) (Nrf2/HO-1) pathway, and inhibited NF-κB and ERK1/2 in a dose-dependent manner The neuroprotective and anti-inflammatory effects of fraxin were also diminished by treatment with GW9662 which is a PPAR-γ antagonist.30,31
In LPS-induced ARDS in mice, fraxin reduced the production of TNF-α, IL-1β, IL-6, Reactive oxygen species (ROS), and Malondialdehyde (MDA), increased superoxide dismutase (SOD), and suppressed NF-κB and MAPK signaling.32
Li et al., 2019 discovered that pretreatment with fraxin decreased protein expressions of NF-κB and nucleotide-binding domain, leucine-rich–containing family, pyrin domain–containing-3 (NLRP3) activated in response to lipopolysaccharide (LPS).33 Aesculin, a hydroxycoumarin, is the 6-O-beta-D-glucoside of esculetin, another organic compound isolated from Cortex fraxini that is structurally related to fraxin. Furthermore, studies in the peritoneum and macrophages demonstrated that aesculin inhibits the production of inflammatory mediators such as iNOS, IL-1, and TNF-α via the PPAR-γ/NF-κB pathway.34
While quercetin inhibits liver inflammation mainly through NF-κB/TLR/NLRP3, it also inhibits LPS-stimulated NO increase by suppressing iNOS.35,36 In addition, in the differentiated human acute monocyte leukemia cell line (THP-1), quercetin might lower cholesterol levels in macrophages with elevated PPAR-γ expression. Quercetin metabolites, such as quercetin-3-glucuronide (Q3G) and quercetin-3′-sulfate, also upregulated PPAR-γ in A549 lung cancer cells.37,38
Flavonoids, such as quercetin and kaempferol, increase PPAR-γ-mediated gene expression through a mechanism distinct from conventional PPAR-γ agonists.39
Dihydroquercetin activates AMPK/Nrf2/HO-1 signaling in macrophages, which mediates its anti-inflammatory effects.40
Our results were consistent with previous studies supporting that fraxin and quercetin upregulated PPAR-γ expression and downregulated TLR-4, stimulated by LPS treatment in macrophage RAW 264.7 cells.
Fraxin + quercetin showed synergistic activity when combined, which may be due to multiple targets involved when coming to their anti-inflammatory mechanism, resulting in suppression of proinflammatory mediators IL-1, IL-6, TNF-α, and suppression of other pathways like iNOS, COX-2, Nrf2/HO-1, NF-κB, NLRP3, TLR-4, and upregulation of PPAR-γ.
A study described the synergistic combination of two bioflavonoids: quercetin and catechin; this combination caused inhibition of the LPS-activated upregulation of iNOS and COX-2.26 Previous studies have shown that drug combinations, especially in phytopharmaceuticals, may activate entirely different sets of genes than those started by each drug alone.41 This may provide another theoretical explanation for the synergistic activity between fraxin and quercetin, despite the differences in their chemical structures.
Our study showed that fraxin, quercetin, and their combination exert anti-cytokine storm activity on LPS-induced RAW246.7 cells by targeting multiple signaling pathways and suppressing TLR-mediated NF-κB. Upregulation of PPAR-γ mediated gene expression (Figure 4A and B) may serve as a foundation for future research into other combinations of fraxin and quercetin and pathways involved in their molecular mechanisms explaining the synergistic cytokine release inhibitory activity. As the current study demonstrates an effect against cytokine release with potential synergism, these outcomes need further exploration using in vivo models of LPS to validate the synergism through pharmacokinetic studies and explore other signaling pathways to explain and relate to the current outcomes.
Zenodo: Anti-cytokine storm activity of fraxin and quercetin, alone and in combination, and their possible molecular mechanisms via TLR4 and PPARγ signaling pathways in LPS-induced RAW 264.7 cell line article data https://doi.org/10.5281/zenodo.7822393.42
This project contains the following underlying data:
• Article data.xlsx (Anti-cytokine storm activity of fraxin and quercetin, alone and in combination, and their possible molecular mechanisms via TLR4 and PPARγ signaling pathways in LPS-induced RAW 264.7 cell line article data).
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Pharmacology, Therapuetics, Toxicology, clinical toxicology and pharmacognosy.
Is the work clearly and accurately presented and does it cite the current literature?
No
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Please see my comments to authors.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
No source data required
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Micobiology and Immunology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 10 Sep 25 |
read | ||
|
Version 1 08 Sep 23 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)