Keywords
Lapatinib resistance, oncology, anti-cancer agents, HER2-amplified cancer, Compensatory pathways, SKBR3 cell line, BT-474 cell line, breast cancer.
This article is included in the Genomics and Genetics gateway.
This article is included in the Oncology gateway.
Lapatinib resistance, oncology, anti-cancer agents, HER2-amplified cancer, Compensatory pathways, SKBR3 cell line, BT-474 cell line, breast cancer.
The human epidermal growth factor receptor 2 (HER2) is a tyrosine kinase receptor that is overexpressed in 20-30% of breast cancers.1 Overexpression of HER2 has been linked with high tumor invasiveness, poor patient prognosis, and low survival rates.2
Since the initial clinical trials of anti-HER2-targeted therapy, targeted cancer therapy has changed the landscape of cancer treatment with high response rates, and several pharmacological drugs have been approved with widespread clinical benefits for patients with metastatic breast cancer. Pharmacological drugs directed against HER2 activity could be classified into monoclonal antibodies (mAb), such as trastuzumab, the gold standard mAb for HER2-positive breast cancer,3 and small-molecule drugs, such as lapatinib.4
Lapatinib is a small molecule drug that binds reversibly to the intracellular kinase domain of HER2 receptors4 and is known as a dual kinase inhibitor which was approved by the US Food and Drug Administration (FDA) in 2007. It is widely used in HER2-positive metastatic breast cancer in combination with capecitabine, trastuzumab, or aromatase inhibitors. Nonetheless, despite the encouraging initial outcomes of targeted therapies, breast cancers often develop resistance to these drugs.5
The mechanisms of acquired resistance to lapatinib have been studied in various HER2-positive cell lines and animal models, and various underlying mechanisms have been suggested including a) the activation of alternative kinases and non-kinases cellular pathways to maintain growth and survival, b) the alteration in the upstream and downstream signalling networks effectors, which are acquired mutation in HER2 gene, and c) the intracellular metabolic changes.6–8
Two independent studies on two HER2-positive cell lines, SKBR39 and BT-474,7 elaborated on the differential underlining resistance gene networks. Komurov and co-workers correlated lapatinib toxicity and acquired resistance with the increase of expression of the glucose deprivation or hypoglycemic response pathways.9 In contrast, Liu et al. reported the overexpression of AXL, a tyrosine kinase receptor, in resistant HER2-positive breast cancer cells.7
Previous studies attempted to explain the basis of acquired resistance to lapatinib based on studying one type of HER2-positive cell lines, and therefore couldn’t provide a comprehensive mechanistic insight derived from multiple cancer cell lines. Herein, we suggest and prove that the systematic analysis of gene expression data in multiple HER-2 positive cell lines, coupled with functional genomics, is more capable of elaborating the network biology of lapatinib’s resistance to enable the prioritization of effective intervention measures. Our studies uncovered the most perturbed oncogenic pathways in two different HER2-positive breast cancer cell lines. Our findings fulfilled the main objective of this work by identifying reliable gene networks that provided an in-depth understanding of the underlying resistance mechanisms to lapatinib in HER2 breast cancers. Our network biology findings led to evidence-based hypotheses regarding promising intervention measures to overcome lapatinib resistance and achieve optimal therapeutic outcomes.
Two microarray data sets, GSE38376 and GSE16179, were obtained from NCBI’s GEO database. Data set GSE38376 profiled gene expression in response to lapatinib treatment in a HER2-positive breast cancer cell lines SKBR3 (i.e., original lapatinib sensitive cells and SKBR3-R (i.e., lapatinib resistance cells).9
The second dataset, GSE16179, profiled gene expression in BT-474 cell lines, as additional examples of HER2-positive and lapatinib-sensitive, and HER2-positive and lapatinib-resistant cell lines, subsequently.7
GEO2R is a web-based tool for microarray data analysis, available from the Gene Expression Omnibus, was used to identify differentially expressed genes (DEGs) across experimental conditions. GEO2R is powered by R’s Bioconductor. We used R 3.2.2, Biobase 2.30.0, GEOquery 2.40.0, and limma 3.26.8.
The pathway analysis module from IPA (Qiagen IPA)10,11 was used to analyze the lists of DEGs in lapatinib respondent/resistant. Gene identifiers and corresponding gene expression levels were uploaded to IPA. The differentially expressed data, which comprised biological processes, canonical pathways, upstream transcriptional regulators, and gene networks, were interpreted using the software's “core analysis” tool.
Using IPA, genes were considered differentially expressed when the adjusted p-value was 0.05 or less with an absolute fold change of ±2 or greater.
Our results led to the prioritization of 859 DEGs (462 upregulated genes and 397 downregulated genes) between the parental and resistant cell lines for GSE38376. Additionally, 1701 DEGs (924 upregulated genes and 777 downregulated genes) for GSE16179. All identified DEGs (p<0.05 and with exact absolute fold-change ≤ -2.5 and ≥ 1.5).
Graphical summary
The graphical summary in our IPA analysis aims to provide an overview of the biological themes of all DEG.
The graphical summary of the gene sets GSE38376 demonstrated a complex interplay between HIF-1α, epidermal growth factor (EGF), and inflammatory factors (TNF and IL1β) pathways (Figure 1). EGF, TNF, and IL1β were predicted to activate homing of the cell and ultimately activate the HIF-1α pathway (Figure 1). On the other hand, the graphical summary of the gene sets GSE16179 demonstrated a complex interplay between β-catenin, Wnt pathways, forkhead box M1 (FOXM1) transcriptional factor, CDK5, and other factors. These pathways are directly related to tumor cell proliferation, movement, and migration (Figure 2).
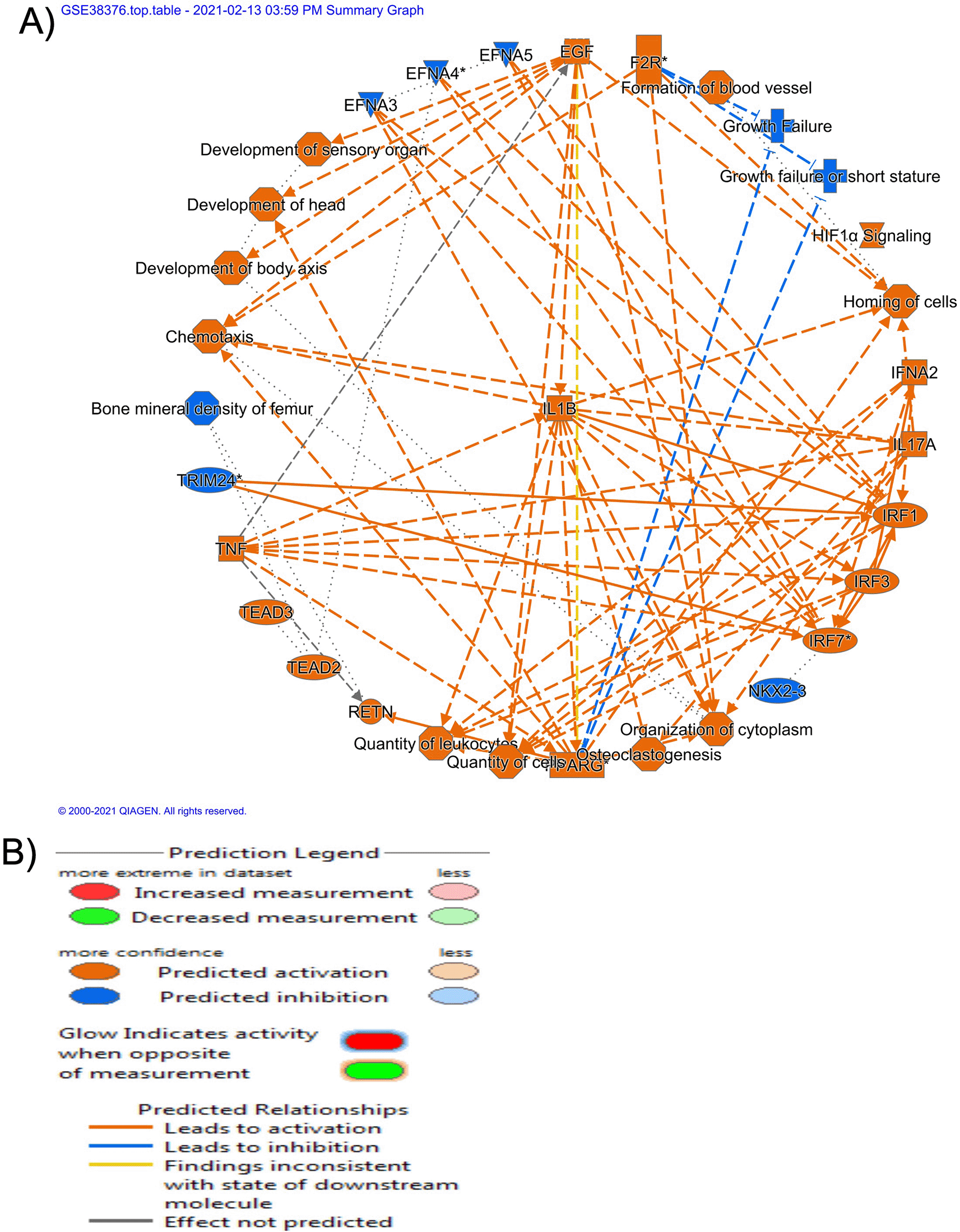
(B) Diverse shapes indicate the protein's various molecular classes. Blue and orange represent inhibition and respectively, activation, whereas red and green represent down- and up-regulation, respectively. A direct contact is indicated by a solid line, an indirect interaction by a dashed line, and an inferred correlation through machine-based learning is indicated by a dotted line. Multiple identifiers in the dataset file that link to a single gene or substance in the Global Molecular Network are denoted by an asterisk (*).
The most significant canonical pathways, enriched by the DEGs, in resistant SKBR3 cell lines were HIF-1α and dopamine degradation pathways. HIF-1α was identified as a canonical pathway, whereas several downstream mediators of both RAS/Raf/MEK and PI3K/AKT/mTOR pathways were predicted to be activated mainly by phosphorylation, notably MEK, mTOR, and RPS6 (Figure 3). The associated HIF-1α and hsp90 were subsequently translocated to nuclease. After that, many cellular processes would be activated, including cellular migration, invasion, survival, glucose uptake, and angiogenesis (Figure 3).
On the other hand, in BT-474 cell lines, Wnt/β-catenin was found to be the top canonical pathway. As shown in Figure 4, the binding of Wnt ligands to the frizzled receptors triggered the activation of downstream effectors, mainly GPB receptors, which subsequently phosphorylated by kinases, such as CK1α and GSK-3β. Ultimately, free cytoplasmic β-catenin will be translocated into the nucleus, displaces TLE/Groucho repressors, and associate with TCF/LEF proteins. This complex transcriptionally recruits Wnt target genes, such as c-Myc and other genes associated with proliferation, including genes (e.g., CD44) associated with cancer stem cell biomarkers. Besides, the complex activates other genes, c-Jun, which triggers cellular adhesion and migration (Figure 4).
Moreover, one of the main pathways found to be downregulated in resistant SKBR3 cell lines was the dopamine degradation pathway with a negative z-score of 1.414.
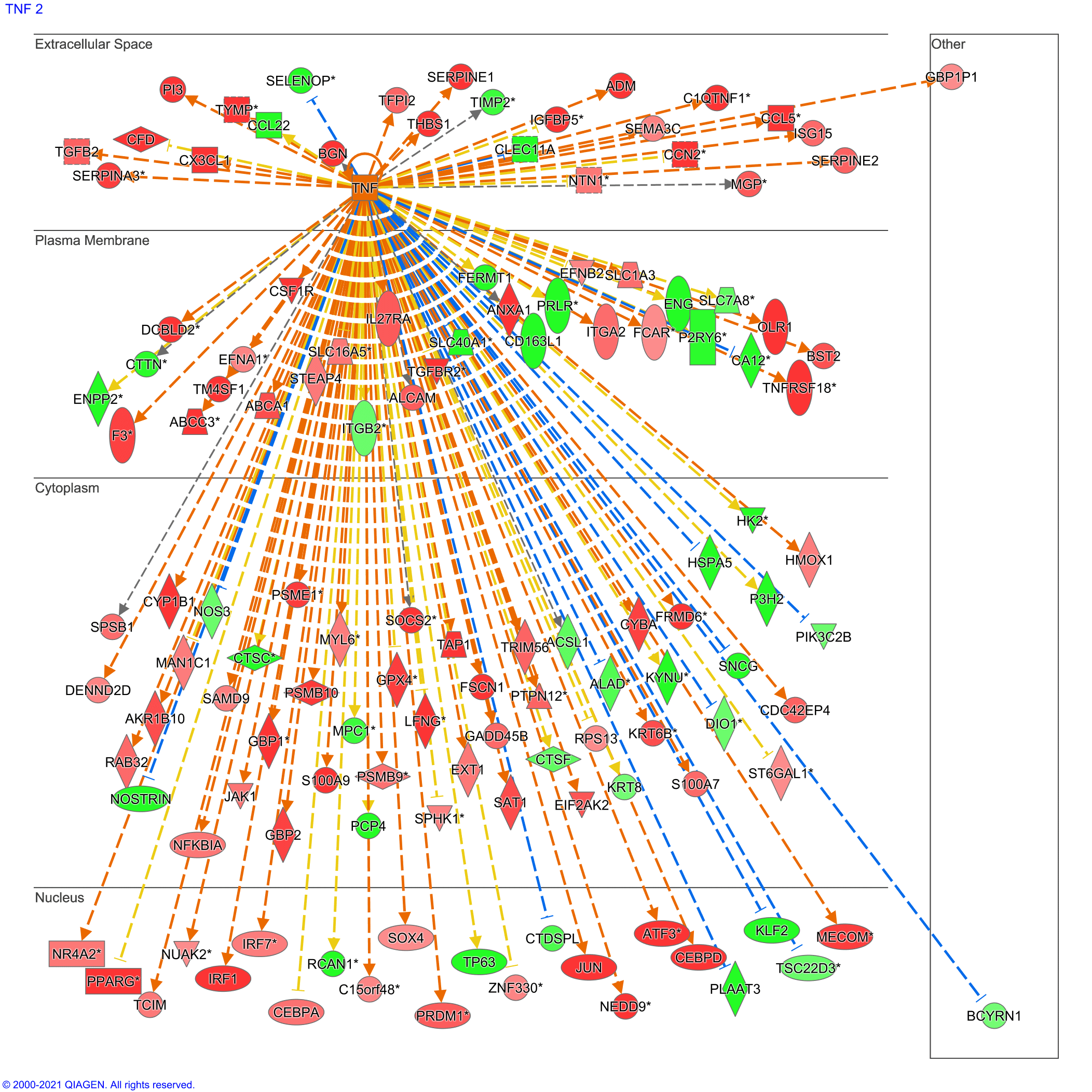
TNF cytokine was predicted to be the central upstream regulator. TNF is regulated and implicated in many mediators and effectors at extracellular, cytoplasmic, and nuclear levels. For instance, 20 genes were predicted to be significantly under-expressed in the extracellular space versus five overexpressed genes (Figure 5).
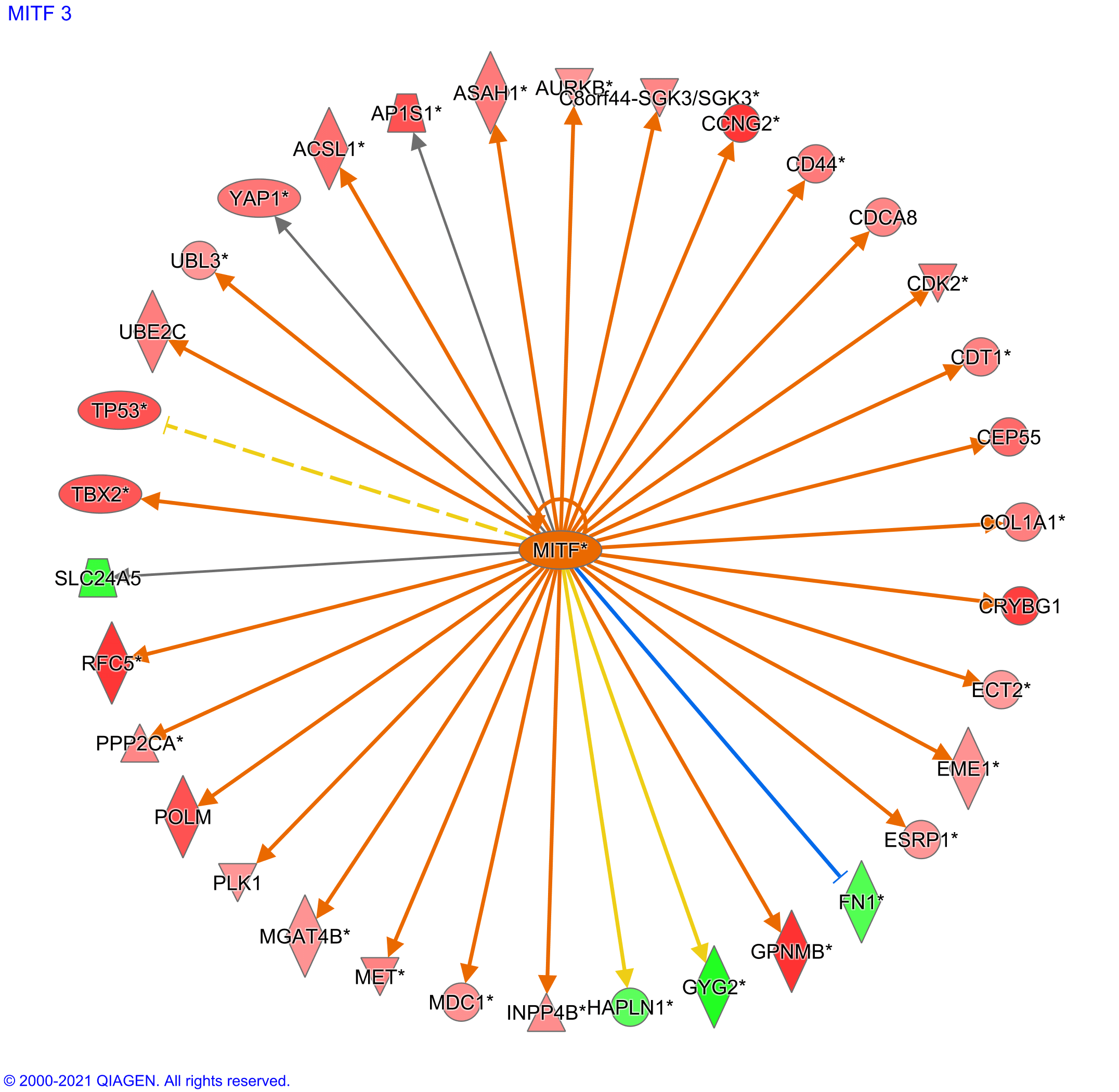
MITF transcription factor was predicted to be the top upstream regulator with a z-score of 4.5 (Figure 6). The MITF transcription factor regulated mainly the overexpression of various mediators and effectors. For instance, 30 genes were predicted to be significantly overexpressed versus four under-expressed genes.
Lastly, both gene datasets were overlayed to predict common pathways or mediators between the two HER2 cell lines. Of the 201 genes related to the top canonical pathway, only 22 genes were predicted to be shared between both resistant cell lines (Table 1).
Several mechanisms have been reported for resistance to lapatinib. Among these are increasing gene expression,12 recruitment of compensatory pathways, most notably HER3 co-receptor, and mutation within the intracellular domain tyrosine kinase domain.13
Although both cell lines are classified as HER2-positive, the discrepancy between the two cell lines might be the critical driving factor for the acquired resistance to lapatinib treatment. The SKBR3 cell line is HER2-positive and derived from adenocarcinoma, while BT-474 cells are HER2-, ER-, and PR-positive and classified as luminal B and derived from invasive ductal carcinoma.14 SKBR3 and BT-474 cell lines reflect two different breast cancer subtypes, and their prognosis and aggressiveness vary significantly. BT-474 reflect luminal B subtypes and they are considered relatively less aggressive than the other type, i.e.HER2-positive subtype (SKBR3). And therefore, BT-474 reflects a more aggressive cancer subtype with a poor prognosis.14
Herein, the HIF-1α signaling pathway was predicted to be the top canonical pathway in the resistant SKBR3 cell line. On the other hand, Wnt/β-catenin was predicted to be the top canonical pathway involved in cellular proliferation and invasion in the resistant BT-474 cell line. Despite the difference, both canonical pathways are directly linked to tumor invasion, where HIF-1α is associated with angiogenesis,15 paving the way for invasion and metastasis. Likewise, the Wnt/β-catenin pathway is directly implicated in tumor epithelial-to-mesenchymal transition (EMT), stemness, invasion, and metastasis.16,17 Therefore, we propose that by switching on invasion and metastasis processes, both cancer cells become chemoresistant to the targeted therapy but with different driving mediators and factors.
Herein, and as shown previously, HIF-1α was predicted to be the top canonical pathway in resistant SKBR3 cell lines. Since HIF-1α is the principal regulator of oxygen homeostasis,18 the upregulation of HIF-1 level, particularly under hypoxic conditions, causes alterations in mitochondrial oxidative metabolism, glucose uptake, energy production, and angiogenesis which ultimately render the cancer cell prone to proliferate, migrate, and survive.15 Evidence from the literature indicated that glucose-deprivation pathways are upregulated in lapatinib-resistant cells.9 Crosstalk between HIF-1α and glucose regulations proteins, including glucose transporters, has been outlined previously.19 Therefore, we propose that cancer-resistant cells could mediate resistance by switching on the HIF-1α pathway, thereby modulating cellular growth under hypoxic conditions.
In breast cancers, the HIFs pathway is a marker of poor prognosis, metastasis promoter, and drug resistance.20 HIF-1α is a chief player in developing drug resistance in breast cancer, including endocrine-based therapy, cytotoxic drugs, and monoclonal antibodies, including 5-fluorouracil, doxorubicin, sorafenib, dasatinib, and trastuzumab. In part, the latter is due to the complex role of HIF-1α in regulating a plethora of cellular activities and functions essential for cell survival.20
As shown in Figure 3, the complex of HIF-1α with Hsp90 was directly involved in the downstream signalling pathways related to cellular survival. Hsp90 binds to HIF-1α and directly induces conformational changes in HIF-1α to initiate its transactivation. In addition, Hsp90 can stabilize HIF-1α against degradation.21 Thus, on the whole, previous results are in concordance with the gene analysis conducted in our study, where we propose that the variously reported resistance mechanism converges at the induction and stabilizing of the HIF-1α transcriptional factor.
Apart from oxygen tension, nutrients, growth factors, and cytokines are well-known regulators of the HIF-1α factor. The complex interaction between the pro-inflammatory cytokines (TNF and IL1β), homing, and invasiveness of malignancies has been intensively reported.22 Moreover, TNF contributes to developing acquired drug resistance, particularly in breast cancer. Furthermore, as outlined above, HIF-1α is mainly linked to invasiveness and metastasis. Therefore, up-regulation of inflammatory cytokines could induce the HIF-1α signalling pathway. Inflammation and TNF-related induction of the HIF-1α is mainly mediated through the NF-κB signalling cascade.23 Consistent with the previous findings, TNF was predicted as the top regulatory gene in resistant SKBR3 cell lines, which could be directly linked with the induction of angiogenesis through the recruitment of the HIF-1α pathway.
As for the GSE16179 gene set, the Wnt/β-catenin signaling pathway was predicted to be the top canonical pathway in resistant BT-474 cell lines. The Wnt/β-catenin signaling pathway involves various physiological processes, including cellular proliferation, differentiation, apoptosis, invasion, and migration.24 Increasing evidence highlighted a dysregulation of the Wnt gene pathway with the evasion of anti-tumor immune attack, alteration of cell metabolism, and cellular invasiveness and mobility in breast cancer.25 Furthermore, the Wnt signalling pathway activates the expression of different genes that regulate cancer stem cells’ phenotype, including self-renewal, migration, and pluripotency. In addition, the β-catenin transcriptional complex is implicated in developing drug resistance and clinical relapse.26,27
Moreover, the crosstalk between PI3K/AKT and Wnt/β-catenin pathways was confirmed previously, and both pathways are associated with poor prognosis and chemoresistance to cancer therapy.28 Targeted agents against various effectors of the Wnt/β-catenin signalling pathway are currently under investigation and exhibited encouraging therapeutic potential at preclinical and clinical trial levels of some malignancies.29
Previously, it was found that resistant HER2-positive breast cancer cells exhibited high levels of the WNT3 ligand of the Wnt/β-catenin pathway, which promoted the EMT-like transition.30 Similarly, Wang and co-workers have elucidated the link between the Wnt/β-catenin signalling pathway and resistance to lapatinib. Wang et al. restored the activity of lapatinib in lapatinib-resistant cells by knocking down β-catenin.31
Based on the comprehensive analysis of the biological network leading to resistance to lapatinib, HIF-1a targeting drugs, such as alvocidib, rapamycin, and cetuximab are predicted to be more effective towards HER2 positive breast cancer patients treated with lapatinib.32 Generally, drugs that interfere with the transcriptional and translational initiation of HIF-1α are predicted to be more effective in combination with the lapatinib HER2 positive subtype.
For luminal B subtype breast cancer patients, it was demonstrated that the Wnt/β-catenin signalling pathway was the main activated down dream pathway in resistant BT-474 cell lines. Therefore, drugs targeting this pathway should be explored in combination with lapatinib to overcome cancer resistance. As outlined earlier, there are many potential drugs currently evaluated for targeting the Wnt/β-catenin pathway, such as cardamonin.29,33
Gene analysis data represent the concordance with the major implicated pathways in targeted drug resistance, including HIF-1α, Wnt/β-catenin, and HER2 pathways. However, to date, the lack of effective preventive strategy to overcome lapatinib resistance spotlights the complex crosstalk between the pathways, mediators, and effectors, and other overlooked ones. Our results highlight the importance of re-classifying HER-positive cancer based on the other essential underlying oncogenic pathways. Due to the discrepancy in resistance mechanisms between the two different HER2-positive cell lines, a global understanding of the significant hallmarks, particularly invasion and metastasis, is warranted before suggesting a combinational therapy with lapatinib or similarly targeted cancer therapy.
Fighsare: supplementary data 1.xlsx, https://doi.org/10.6084/m9.figshare.21970766.v1. 32
Fighsare: supplementary data 2.xlsx, https://doi.org/10.6084/m9.figshare.21970853.v1. 33
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
We would like to offer our heartfelt gratitude to the Faculty of Pharmacy and Medical Sciences at the University of Petra for providing us with the chance to complete this research. The authors would also like to thank the Deanship of Scientific Research and Graduate Studies for their invaluable assistance.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Mechanisms of resistance to radiotherapy and chemotherapy in breast Cancer
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |
|---|---|
| 1 | |
|
Version 1 09 Mar 23 |
read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)