Keywords
Fin clipping, skin swabbing, lidocaine, analgesia, zebrafish, stickleback
This article is included in the NC3Rs gateway.
This article is included in the Cell & Molecular Biology gateway.
Fin clipping, skin swabbing, lidocaine, analgesia, zebrafish, stickleback
Scientific benefit(s)
3Rs benefit(s)
• Skin swabbing is a refined technique to collect DNA compared to fin clipping, causing fewer health and welfare changes.
• It may be possible to use fewer animals in subsequent experiments when collecting DNA by skin swabbing rather than fin clipping.
Practical benefit(s)
• Skin swabbing is quick to perform and gives reliable results across different institutions.
• There is no need to use anaesthetic, analgesic, or sharp blades when skin swabbing.
Current applications
• The current applications of skin swabbing are to collect DNA samples from small-bodied fish species for use in PCR amplification or genotyping.
Potential applications
Refining the experimental techniques applied to laboratory animals is a vital part of scientific research. Experimental fish are often identified by collecting a small sample of DNA that can be used for genotyping. There are several procedures that can be used to collect DNA from fish including fin clipping and skin swabbing. Fin clipping is the more common technique, and it involves anaesthetising a fish and removing a small piece of fin tissue before allowing it to recover in fresh system water. However, there is evidence showing that fin clipping is harmful for fish, indicated by stress axis activation, changes in behaviour and negative effects on survival and growth.1–6
As an alternative to fin clipping, our recent research has investigated the welfare benefits of using skin swabbing to collect mucus samples from small fish species.7–9 The swabbing procedure involves restraining a non-anaesthetised fish upon a wetted sponge. A mucus sample is collected from the fish’s flank using a rayon-tipped swab. Skin swabbing has been shown to be less invasive than fin clipping since it activates fewer endocrine, genetic and behavioural indicators of stress and leads to less variability in post-hoc data collection.7,8 In addition, the skin swabbing protocol is easy to perform and leads to reliable DNA sampling.8
In this study we have investigated ways to improve the skin swabbing protocol. We first independently verified our study comparing changes to cortisol release and behaviour following fin clipping or skin swabbing using fish held at the University of Gothenburg. In addition, we examined whether swabbing could be improved by applying the analgesic lidocaine. Previous research suggests that analgesia administration post-fin clipping improves fish welfare.3,4 We found no difference in the impact of swabbing when carried out with or without analgesia. However, although lidocaine significantly improved the welfare of fin-clipped fish, this procedure brings additional steps to the fin clipping protocol, and lidocaine can affect fish for significant periods of time. Our results confirm that swabbing is a refined technique to collect DNA from small fish species.
All work was conducted under a UK Home Office licence (University of Leicester, no. P8F9CCE8B) or a local ethics permit (University of Gothenburg, no. 5.8.18-16941/2021). Zebrafish and sticklebacks were kept under standard conditions in accordance with institutional guidelines for animal welfare. Three-spined sticklebacks were generated by in vitro fertilisation as described in.10 They had an average length of 39.02 ± 5.81 mm and an average weight of 0.83 ± 0.41 g. The water parameters were pH ~7.1, 0 ppm ammonia, 0 ppm nitrate, ~4 ppm nitrite and ~4000 μs/cm conductivity. AB wild-type zebrafish kept at the University of Leicester were generated by in-crossing parental stock. Zebrafish used in the lidocaine experiment had an average length of 29.67 ± 2.38 mm and an average weight of 0.24 g ± 0.15 g. Those used in the water supplements experiment had an average length of 41.13 ± 3.79 mm and an average weight of 0.94 ± 0.52 g. The water parameters were pH ~7.1, 0 ppm ammonia, 4 ppm nitrate, ~0 ppm nitrite and ~525 μs/cm conductivity. AB wild-type zebrafish kept at the University of Gothenburg were obtained from the Zebrafish Core Facility, Karolinska Institute, Stockholm, Sweden. They had an average length of 25.86 ± 1.02 mm and an average weight of 0.22 g ± 0.12 g. The water quality parameters were pH ~7.4, <0.1 ppm ammonia, <10 ppm nitrate, and <0.1 ppm nitrite. No fish of either species died during these experiments and all animals were killed by a Schedule 1 procedure at the end of the study.
Experiment 1. Cortisol and measurements of behaviour (University of Gothenburg)
Cortisol release and measurements of behaviour using the Fish Behavioural Index (FBI) were carried out in Gothenburg. A total number of 48 zebrafish were used. Experiments were conducted as outlined in.3 Zebrafish were randomly selected and transferred to a semi-closed recirculation system containing two parallel rows of glass tanks (20 × 30 × 20 cm; n = 1 fish per tank). Fish were acclimatised in their individual tanks for two weeks prior to experimentation. They were in visual contact with adjacent tanks until the evening prior to experimentation when two opaque pieces of plastic were placed between tanks to visually isolate individuals. Fish were randomly assigned by haphazard netting from a home tank into one of the following groups: Fin Clip; Fin Clip plus Lidocaine; Skin Swab or Skin Swab plus Lidocaine (n = 12 per group). At the start of the experiment the water flow to the experimental tanks was switched off and 5 mg/L lidocaine was administered to the tank. Behaviour was compared at pre-treatment and 1, 2, 3 and 6 h afterwards. At each time point behaviour was recorded for 25 min using video cameras linked to FBI tracking software. The FBI assigns one value based upon activity and distance travelled to classify behaviour as: Healthy (0.84-1.0); OK (0.67-0.83); Unhealthy (0.33-0.66); or Abnormal (0-0.32) (Deakin et al., 2019). After the video recording, a water sample was obtained for cortisol analysis.1 Water samples were collected into two clean 120 mL containers per treatment group using siphons. Blank water samples (n = 2) were taken directly from empty tanks at each sampling point to account for residual cortisol levels (ranging from 37-56 pg/cartridge) and blank values were subtracted from the corresponding experimental sample.11 Following collection, samples were immediately frozen at -20°C and were allowed to thaw at 4°C for 24 hours before cortisol extraction at a later date. Water samples were filtered with 0.45 micron nitrocellulose filters (10 cm diameter, Whatman, UK) before undergoing solid phase extraction using C-18 Sep-pak cartridges fitted to a mini-pulse machine set at 25 ml min-1. Cartridges were first primed with 5 ml methanol and washed with 5 ml distilled water. The water sample was then passed through the cartridge, followed by a final wash with 5 ml distilled water. All cartridges were labelled and allowed to dry before being frozen for future analysis (-20°C). Cortisol was assayed blind using 200 μL of eluate in a radioimmunoassay.12
Experiment 2. Administration of lidocaine (University of Leicester)
In separate experiments carried out in Leicester, we investigated the welfare impact of administering analgesia prior to DNA sampling and stress relief water supplements post-sampling. Two weeks prior to experimentation, fish were caught and were distributed by blind randomization into ten tanks on the system, (six for analgesia studies and four for water supplements with nine sticklebacks in 13.4 L tanks and nine zebrafish in 3.5 L tanks). The tanks were located in central locations of large racks within the respective species rooms, ensuring that all tanks received similar illumination and were surrounded by other tanks on the sides. No enrichment was provided. The number of fish per group was based on numbers needed for qPCR analysis. On the day of the experiment tanks were removed from the rack and placed on a trolley for 30 minutes before the start. Lidocaine studies were carried out on one day and water supplements on the following day. This was repeated two more times each week to give three technical replicates for each study. On the day of the experiment a 400 mg/L stock solution of lidocaine (Sigma-Aldrich) was prepared and diluted to 2 mg/L4 using reverse osmosis water. Lidocaine was applied by immersion for 45 min before manipulation. Six groups were investigated in both species: Non-manipulated with no pain relief (un-manipulated); Netted, swabbed and released with no pain relief; netted, anaesthetised, fin clipped and allowed to recover with no pain relief; Un-manipulated after immersion in lidocaine; Netted, swabbed and then released after immersion in lidocaine; netted, anaesthetised and fin clipped after immersion in lidocaine. The total number of individuals used were 162 sticklebacks and 162 zebrafish (9 × 6 groups × 3 independent replicates).
For details of DNA collection by skin swabbing or fin clipping, and gene expression analysis refer to.8 The expression of five stress marker genes were investigated: brain-derived neurotrophic factor (bdnf), corticotropin releasing hormone a (crha), corticotropin releasing hormone b (crhb), galanin (galn) and neuropeptide y (npy).8 The data were normalised against the geometric means of ribosomal protein L8 (rpL8), ribosomal protein L13A (rpL13A) and ubiquitin (ubiq) genes in sticklebacks and ribosomal protein L13A (rpL13A) and elongation factor 1a (elf1a) genes in zebrafish. The fold change was calculated using the 2-ddCT method.13 Primer details are shown in Table 1.
Post-hoc Pairwise comparisons of lidocaine treatment and DNA method for each fish type. Degrees of freedom are 175 except for galn where they are 94 as this was not tested using zebrafish samples. 162 sticklebacks and 162 zebrafish (9 x 6 groups x 3 independent replicates) were used and 3 technical replicates were performed in the qPCR.
Statistical analyses were carried out using GraphPad Prism7 and RStudio. Data were tested for normality using the using the Shapiro-Wilk test, followed by a non-parametric Kruskal-Wallis test and Dunn's multiple comparisons test comparing each treatment to the control group. Gene expression data was Log2 transformed. An ANOVA was carried out for each gene separately (model: log2(expression) = lidocaine + treatment + fish + lidocaine:treatment:fish). Post-hoc tests were carried out using the Emmeans package version 1.5.4 in R.14
Our previous research has shown that skin swabbing is a refined technique to collect DNA compared to fin clipping.8 In the first set of experiments we treated 48 zebrafish with the analgesic lidocaine before either fin clipping or skin swabbing. We investigated changes to locomotion and space usage in the tank using the Fish Behavioural Index (FBI), an automated system that gives an unbiased readout of fish welfare.5 Fin clipping caused a sharp decrease in fish welfare, demonstrated by a reduction of FBI units that persisted over time (>6 h). Fin-clipped fish displayed a score of 0.18 FBI units (representing abnormal behaviour) six hours after fin clipping. Pre-treatment with lidocaine prevented this decrease in welfare. However, since FBI scores had not normalised after 6 h, lidocaine may only represent temporary relief for fin-clipped fish. In comparison, skin swabbed animals displayed similar profiles regardless of the treatment group (with or without lidocaine). All swabbed fish displayed ‘healthy’ behaviour profiles (FBI scores 0.90-0.96) (Figure 1B). We next compared excretion of cortisol. In keeping with other published studies,4,8 fin clipping in the absence of lidocaine led to heightened cortisol release compared to fin clipping with lidocaine, or skin swabbing with or without lidocaine. Importantly, there was no significant difference between the amount of cortisol excreted by zebrafish following skin swabbing with or without lidocaine, suggesting that skin swabbing is not as stressful to fish as fin clipping and that it is not refined by analgesia administration (Figure 1A). We were thus able to confirm the findings of our previous research8 using fish housed in a different country and data collected by researchers that were not involved in earlier studies.
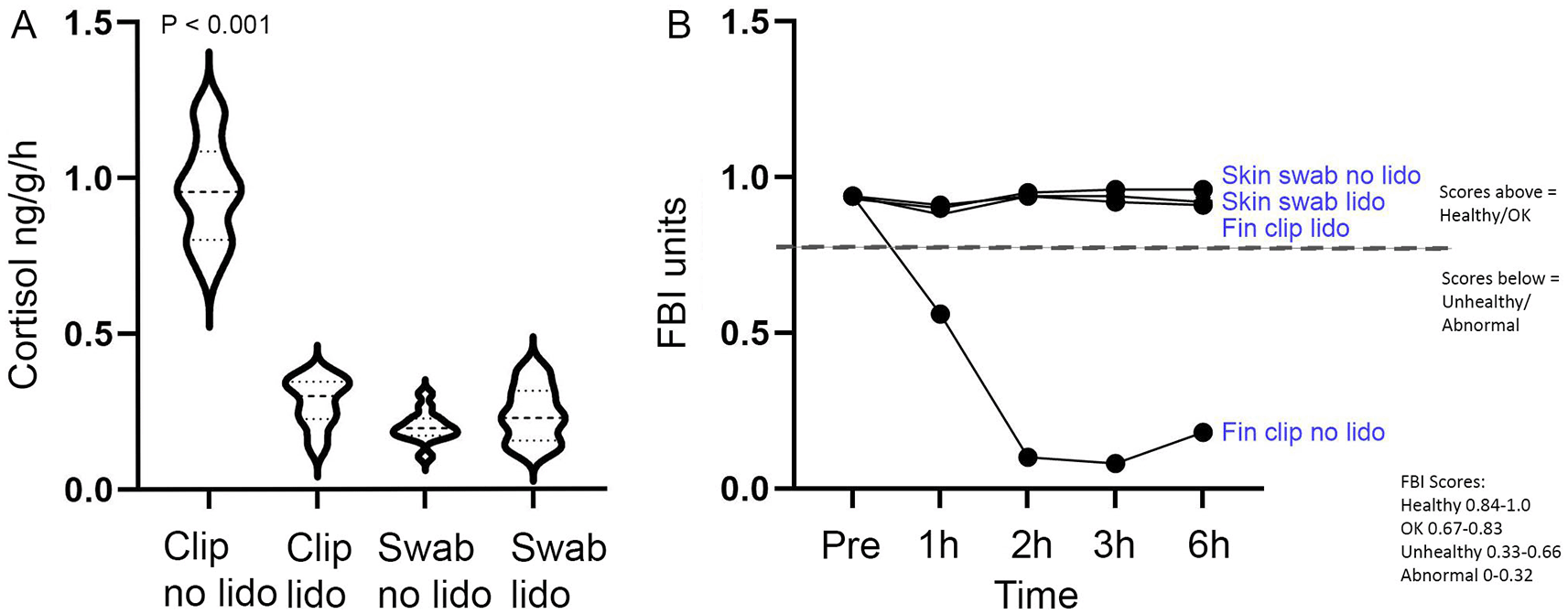
A. There is no significant difference in cortisol release between fin clipping with lidocaine treatment, or skin swabbing with and without lidocaine treatment. Treatment groups: fin clipping without lidocaine (Clip no lido); fin clipping following lidocaine pre-treatment (Clip lido); skin swabbing without lidocaine (Swab no lido and skin swabbing following lidocaine pre-treatment (Swab lido). One-way ANOVA followed by Dunnett’s multiple comparisons test. Clip no lido vs Clip lido, n=12 DF=44, p < 0.001; Clip no lido vs Swab no lido, n=12, DF=44, p < 0.001; Clip no lido vs Swab lido, n=12, DF=44, p < 0.001. B. Fin clipping in the absence of lidocaine decreases zebrafish health status compared to skin swabbing with or without lidocaine treatment. Treatment groups: fin clipping without lidocaine; fin clipping following lidocaine pre-treatment; skin swabbing without lidocaine and skin swabbing following lidocaine pre-treatment. Scores from the FBI are interpreted as follows: Healthy 0.84-1.0, OK 0.67-0.83, Unhealthy 0.33-0.66 and Abnormal 0-0.32.
We next used quantitative PCR to compare the expression level of five genes related to the stress response in sticklebacks and zebrafish: brain derived neurotrophic factor (bdnf); corticotropin releasing hormone a (crha); corticotropin releasing hormone b (crhb); galanin (gln) and neuropeptide y (npy). We compared the effect of lidocaine treatment in fish that were either un-manipulated, fin clipped or skin swabbed either with or without lidocaine (n = 27 for each treatment group, 162 in total for each species). Heightened expression of these genes would indicate higher stress levels in fish. In the absence of lidocaine, fin clipping led to heightened expression of crha, galn and npy compared to un-manipulated sticklebacks (Figure 2). There was also a significant difference in bdnf expression between skin swabbed and fin clipped sticklebacks (Figure 2). Pre-experimental lidocaine treatment heightened npy expression in sticklebacks that were not used for DNA collection, suggesting that lidocaine application can be harmful, or that the concentration and treatment regime needs to be optimised (Figure 2). However, lidocaine treatment decreased the expression of bdnf, crha and galn in fin clipped sticklebacks compared to sticklebacks that had been fin clipped without analgesia (Figure 2). There were no significant differences in gene expression between skin swabbed sticklebacks with or without lidocaine application.
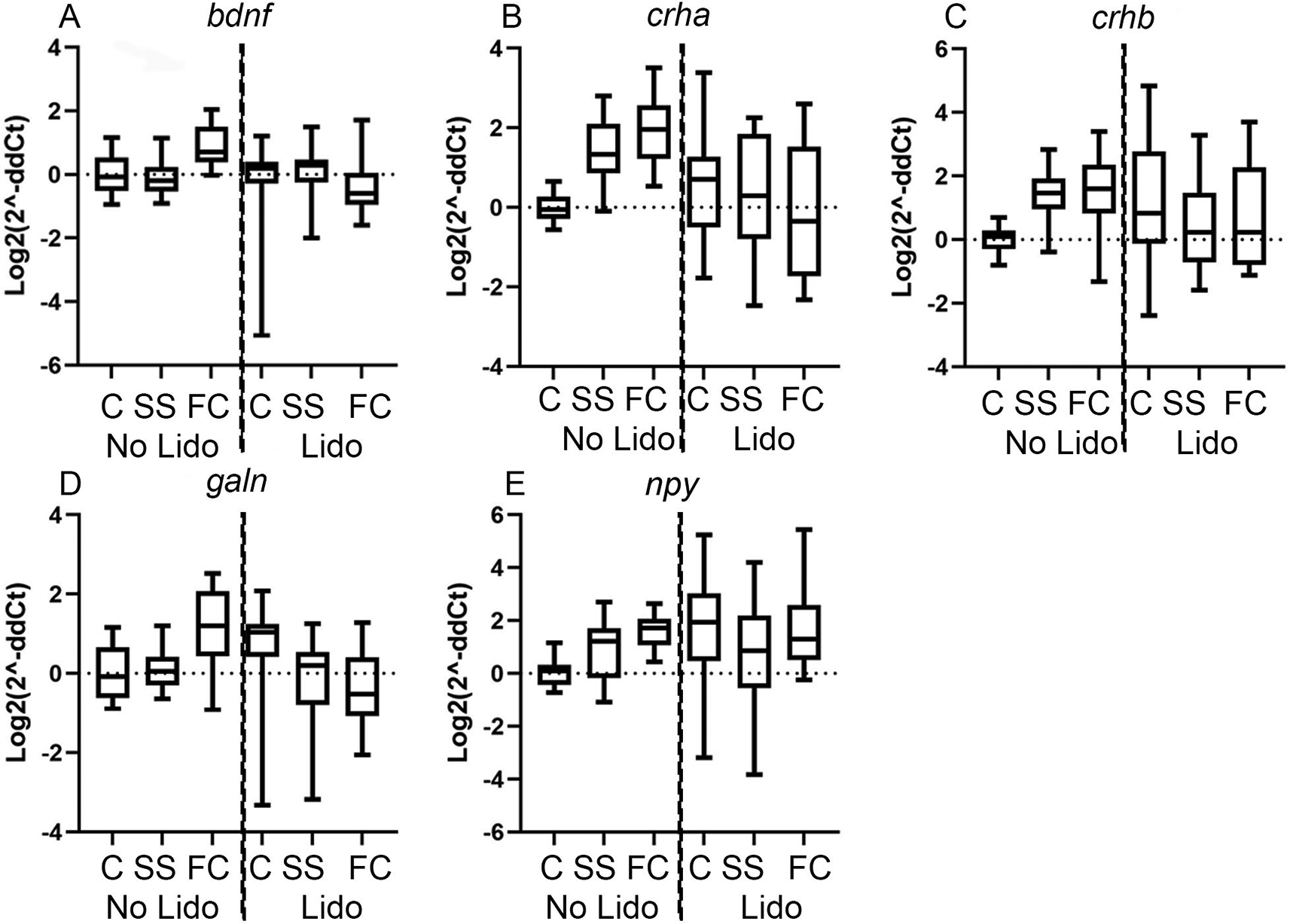
qPCR data showing expression relative to the geometric mean of rpL8, rpL13A and ubiq of (A) stickleback brain-derived neurotrophic factor (bdnf). (B) stickleback corticotropin releasing hormone a (crha). (C) stickleback corticotropin releasing hormone b (crhb). (D) stickleback galanin (galn). (E) stickleback neuropeptide y (npy). Groups include: 1) un-manipulated control fish (control no lidocaine); 2) skin swabbed fish (swab no lidocaine); 3) Fin clipped fish; (clip no lidocaine); 4) un-manipulated fish pre-treated with lidocaine (control lidocaine); 5) lidocaine pre-treated skin swabbed fish (swab lidocaine) and 6) lidocaine pre-treated fin clipped fish (clip lidocaine). For each group n = 27 (162 in total). Each qPCR consisted of three technical replicates. Comparisons include: a) un-manipulated no lidocaine versus fin clip no lidocaine fish (1 versus 3); b) fin clip no lidocaine versus skin swab no lidocaine (3 versus 2); c) un-manipulated no lidocaine versus un-manipulated lidocaine (1 versus 4); d) fin clip no lidocaine versus fin clip with lidocaine (3 versus 6); e) un-manipulated no lidocaine versus swab no lidocaine (1 versus 2) and f) skin swab no lidocaine vs swab with lidocaine (2 versus 5). Primer sequences are included in Table 1 and full statistical information is included in Table 2. Group legend: C = un-manipulated fish, SS = skin swabbed fish, FC = fin clipped fish.
In the absence of lidocaine there was a significant increase in npy expression in the fin clipped zebrafish compared to the skin swabbed group (Figure 3). Application of lidocaine to un-manipulated zebrafish caused a significant decrease in the expression of crha, crhb and npy (Figure 3). These findings are difficult to interpret because it is not clear what a decrease in gene expression represents in terms of fish welfare. Further research comparing gene expression to other welfare readouts is needed to clarify this point. Similarly, lidocaine treatment decreased the expression of bdnf, crha, crhb and npy in fin clipped zebrafish compared to the no-lidocaine group. Finally, lidocaine treatment also decreased bdnf and crhb expression following skin swabbing compared to the swabbing without lidocaine (Figure 3). In summary, skin swabbing is a refined technique to collect DNA, even without lidocaine application.
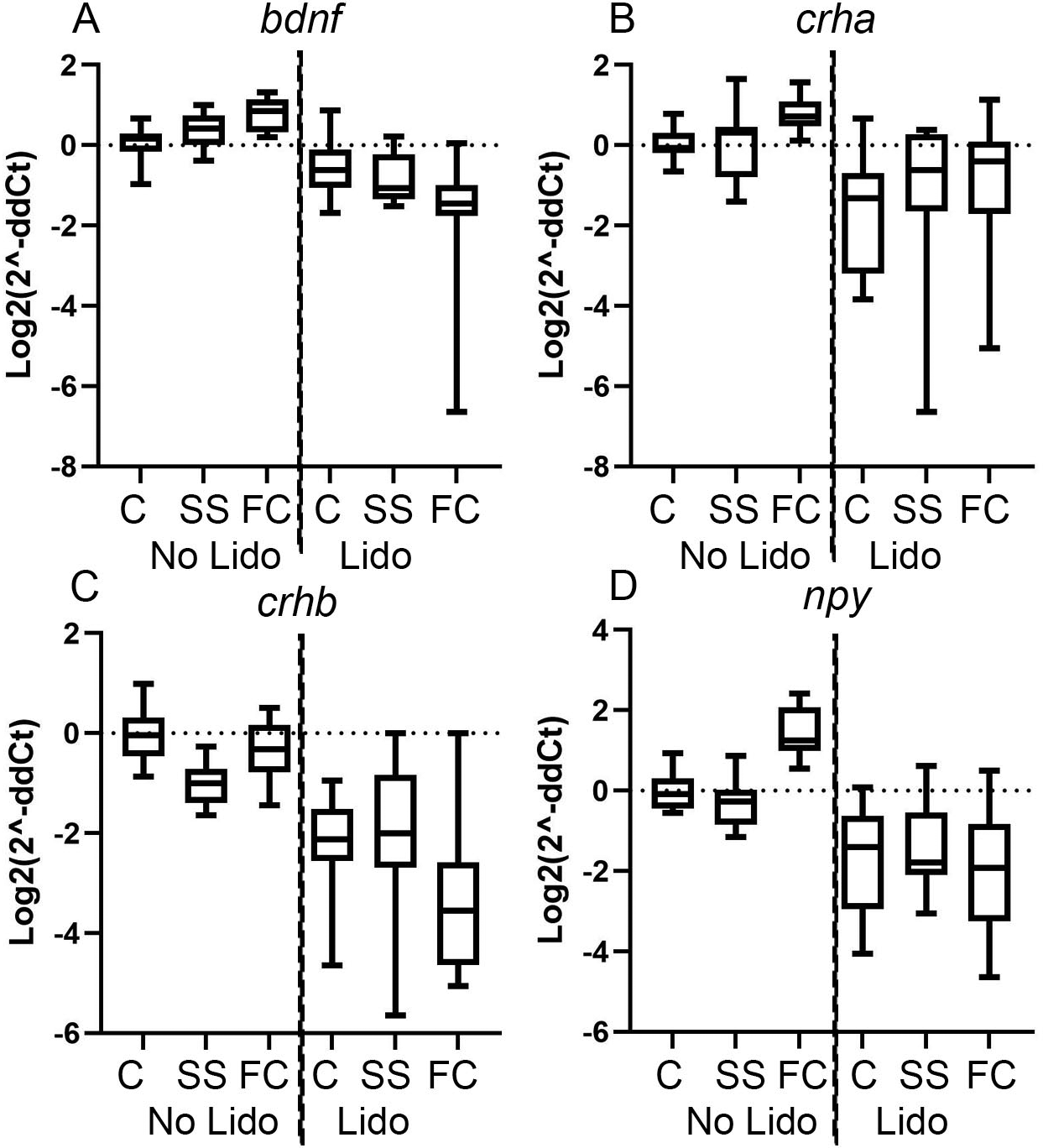
qPCR data showing expression relative to the geometric mean of rpL13A and elf1A of (A) zebrafish brain-derived neurotrophic factor (bdnf). (B) zebrafish corticotropin releasing hormone a (crha). (C) zebrafish corticotropin releasing hormone b (crhb). (D) zebrafish neuropeptide y (npy). Groups include: 1) un-manipulated control fish (control no lidocaine); 2) skin swabbed fish (swab no lidocaine); 3) Fin clipped fish; (clip no lidocaine); 4) un-manipulated fish pre-treated with lidocaine (control lidocaine); 5) lidocaine pre-treated skin swabbed fish (swab lidocaine) and 6) lidocaine pre-treated fin clipped fish (clip lidocaine). For each group n = 27 (162 in total). Each qPCR consisted of 3 technical replicates. Comparisons include: a) un-manipulated no lidocaine vs fin clip no lidocaine fish (1 versus 3); b) fin clip no lidocaine versus skin swab no lidocaine (3 versus 2); c) un-manipulated no lidocaine versus un-manipulated lidocaine (1 versus 4); d) fin clip no lidocaine versus fin clip with lidocaine (3 versus 6). e) un-manipulated no lidocaine versus swab no lidocaine (1 versus 2) and f) skin swab no lidocaine versus swab with lidocaine (2 versus 5). Primer sequences are included in Table 1 and full statistical information is included in Table 2. Group legend: C = un-manipulated fish, SS = skin swabbed fish, FC = fin clipped fish.
We have replicated our findings from an independent study in another facility, using different fish, equipment and researchers, demonstrating that swabbing is a refinement from fin clipping. We also confirmed that lidocaine is an effective analgesic for diminishing pain caused by the fin clipping procedure. Therefore, it is recommended that lidocaine should be provided when fin clipping is used. However, the changes in gene expression in fish exposed to lidocaine with no additional manipulation show that administration of analgesia is a procedure that can result in complex responses, including potential aversion. In addition, it is possible that longer-term welfare impacts remain once the lidocaine wears off, meaning that lidocaine treatment is not ideal. Swabbing is not further refined by administering lidocaine, suggesting swabbing does not cause discomfort. As swabbing is a refinement even in the absence of analgesia, the use of controlled substances can be avoided. We have therefore demonstrated that swabbing represents a true refinement to fin clipping for DNA collection.
Open Science Framework: Lidocaine and water conditioners, https://doi.org/10.17605/OSF.IO/6MD5V. 15
This project contains the following underlying data:
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
Figshare: ARIVE Essential 10 guidelines for “Validating skin swabbing as a refined technique to collect DNA from small-bodied fish species”, https://doi.org/10.6084/m9.figshare.21603024.v1. 16
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Are the 3Rs implications of the work described accurately?
Yes
Are a suitable application and appropriate end-users identified?
Yes
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Behavioral neurobiology of fish (molecular biology, histology, behavior, stress and welfare)
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Developmental biology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 09 Jan 23 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
Please enter your details to receive updates from the NC3Rs
We'll update you about the NC3Rs and the NC3Rs Gateway.
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)