Keywords
Caretta caretta, Loggerhead sea turtle, genome sequence, chromosomal, reptile
This article is included in the Genomics and Genetics gateway.
This article is included in the Nanopore Analysis gateway.
Caretta caretta, Loggerhead sea turtle, genome sequence, chromosomal, reptile
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Archelosauria; Testudinata; Testudines; Cryptodira; Durocryptodira; Americhelydia; Chelonioidea; Cheloniidae; Caretta; Caretta caretta Linnaeus 1758 (NCBI txid 8467).
The loggerhead sea turtle, Caretta caretta, is one of only seven extant marine turtle species and is globally distributed throughout the subtropical and temperate regions of the Mediterranean Sea and Pacific, Indian and Atlantic Oceans (Wallace et al., 2010, Casale and Tucker, 2015). The species is divided in various Regional Management Units (RMUs) and management units (MUs) that vary greatly by population size, geographic range, and population trends (Wallace et al., 2010, Casale and Tucker, 2015, Shamblin et al., 2014). Events such as fisheries bycatch (Caracappa et al., 2018, Pulcinella et al., 2019), human intrusion and disturbance (Mazaris et al., 2009), oceanic pollution (Savoca et al., 2018), and climate change and severe weather (Alduina et al., 2020) have caused the global population to continuously decline (Casale and Tucker, 2015). Consequently, the highly migratory C. caretta requires the collaborative efforts of numerous international conservation and protection organizations (Species at Risk Act, 2002), and is currently listed as Vulnerable by the International Union for the Conservation of Nature (IUCN) (Casale and Tucker, 2015). The genome of C. caretta was sequenced as part of the Canadian BioGenome Project (CBP) and CanSeq150 initiatives. The C. caretta genome will provide insights into genomic diversity and architecture, and inform conservation genomics applications.
Blood samples from an adult female and a juvenile of unknown sex were collected from the Fondazione Cetacea (43.9940 N, 12.6745 E) by Nicola Ridolfi (veterinarian; Fondazione Cetacea). Animal husbandry and welfare were overseen by Fondazione Cetacea. The specimens were transferred to Canada with two CITES permits between institutions (IT002 and CA027).
High-molecular weight (HMW) DNA was extracted from nucleated blood using the MagAttract HMW DNA kit (QIAGEN, Germantown, MD, USA). Nanopore genome libraries were constructed according to manufacturer instructions and sequenced using the PromethION instrument (Oxford Nanopore Technologies). A PCR-free genome library was sequenced in a multiplexed pool of an Illumina NovaSeq 6000 instrument S4 flowcell with paired-end 150 bp (PE150) reads. A Hi-C library was constructed using the Arima-HiC kit 2.0 (Arima Genomics, San Diego, CA) and the Swift Biosciences Accel-NGS 2S Plus DNA Library Kit (Integrated DNA Technologies, Mississauga, ON, Canada) and subjected to PE150 sequencing on an Illumina NovaSeq 6000 instrument. All lab work were performed at Canada’s Michael Smith Genome Sciences Centre at BC Cancer.
Assembly was carried out using Redbean (Ruan and Li, 2019), followed by four rounds of racon (Vaser et al., 2017) polishing and medaka (medaka, n.d.) polishing. Scaffolding with Hi-C data was carried out using nf-core/hic workflow (Servant and Peltzer, 2019), Salsa (Ghurye et al., 2019) and LongStitch (Coombe et al., 2021). The Hi-C scaffolded assembly was polished using Illumina short-reads using Pilon (Walker et al., 2014). Four rounds of manual assembly curation and re-scaffolding with nf-core/hic workflow (Servant and Peltzer, 2019) and Salsa (Ghurye et al., 2019) corrected 54 missing/misjoins. These changes were visualized using JupiterPlots (Chu, 2018) and Juicer (Durand et al., 2016b). The final sequence was analyzed using BlobToolKit (Challis et al., 2020). Software tools and versions are listed in Table 3.
| Project accession data | |
|---|---|
| Assembly identifier | rCarCar2 |
| Species | Caretta caretta |
| Specimen | SJ_126, SJ_184 |
| NCBI Taxonomy ID | 8467 |
| BioProject | PRJNA826225 |
| BioSample ID | SAMN28968396, SAMN27958248 |
| Isolate Information | SJ_184/204:Loco2, SJ_126:Eziel1 |
| Raw data accessions | |
| Oxford Nanopore PromethION | SRX15677840, SRX15677841 |
| Hi-C Illumina | SRX15677843 |
| Illumina short-read | SRX15677842 |
| Genome assembly | |
| Assembly accession | GCA_023653815.1 |
| Assembly name | GSC_CCare_1.0 |
| Span (Mb) | 2,134 |
| Number of contigs | 2,753 |
| Contig N50 length (Mb) | 18,214 |
| Number of scaffolds | 2,008 |
| Scaffold N50 length (Mb) | 130,956 |
| Longest scaffold (Mb) | 345.7 |
| BUSCO* genome score | C:96.1%[S:95.2%,D:0.9%],F:0.4%,M:3.5%,n:7480 |
The genomes of two unrelated loggerhead sea turtles were sequenced from the same population collected from the Fondazione Cetacea hospital, Riccione, Italy. A total of 39-fold coverage in Nanopore PromethION long reads were generated from a single adult female. Approximately 50-fold coverage in Illumina NovaSeq6000 150 bp paired-end (PE150) reads and 18-fold coverage in Illumina NovaSeq6000 Hi-C sequencing were generated from a second individual. Primary assembly contigs from Nanopore data were further polished with Illumina PE150 shotgun sequencing data and scaffolded with Hi-C data. The final assembly has a total length of 2.13 Gb in 2007 sequence scaffolds with a scaffold N50 of 130.95 Mb (Table 1). The majority (98.0%) of the assembly sequence was assigned to 28 chromosomal-level scaffolds representing the species’ known 28 autosomes (Kamezaki, 1989, Machado et al., 2020) (numbered by sequence length; Figure 1–Figure 4; Table 2). Aligned reads from the second turtle to the final assembly had an estimated heterozygosity of 0.11% (2,449,606 heterozygous hits). Determining gene coverage using BUSCO, we estimated 96.1% gene completeness using the sauropsida_odb10 reference set (Manni et al., 2021). The assembly was compared to a previous chromosome-scale assembly of the closely-related green sea turtle, Chelonia mydas (Wang et al., 2013), which has been reported to hybridize with the loggerhead sea turtle (James et al., 2004, Vilaça et al., 2012). The loggerhead sea turtle assembly showed strong synteny to the green sea turtle assembly, as shown in Figure 5. The primary haplotype (rCheMyd1.pri.v2) of the green sea turtle was downloaded from NCBI on July 16, 2022.
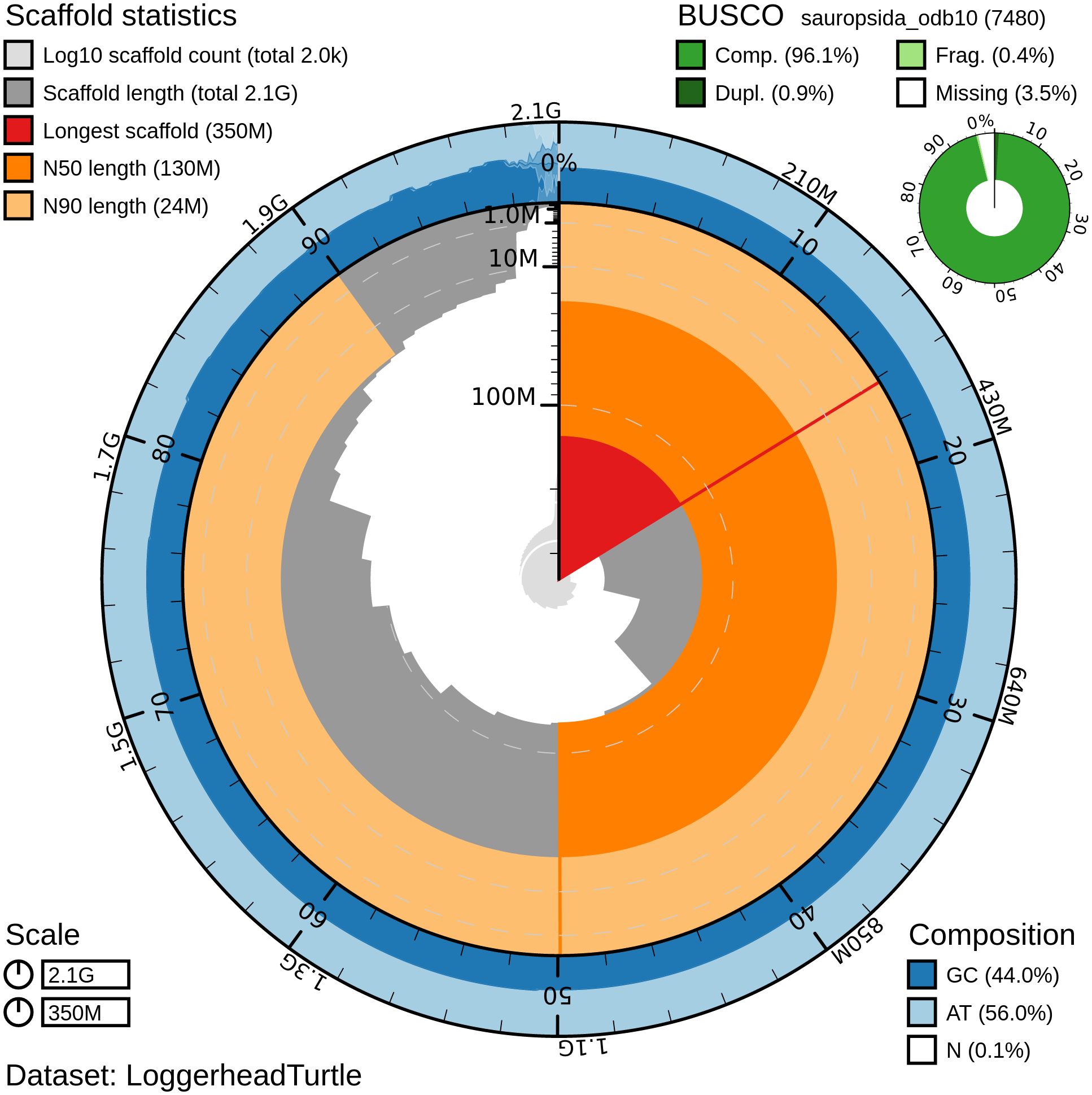
Snail plot showing N50 metrics, base pair composition and BUSCO gene completeness for C. caretta (rCarCar2) generated from Blobtoolkit v.2.6.4 (Challis et al., 2020). The plot is divided into 1,000 size-ordered bins around the circumference with each bin representing 0.1% of the 2,134,012,717 bp assembly. The distribution of chromosome lengths is shown in dark grey with the plot radius scaled to the longest chromosome present in the assembly (345,741,823 bp) shown in red. Orange and pale-orange arcs show the N50 and N90 chromosome lengths (130,956,235 and 23,648,662 bp, respectively). The pale grey spiral shows the cumulative chromosome count on a log scale with white scale lines showing successive orders of magnitude. The blue and pale-blue area around the outside of the plot displays the distribution of GC (blue), AT (pale blue) and N (white) percentages using the same bins as the inner plot. A summary of complete (96.1%), fragmented (0.4%), duplicated (0.9%), and missing (3.5%) BUSCO genes in the sauropsida_odb10 set is show in the top right.
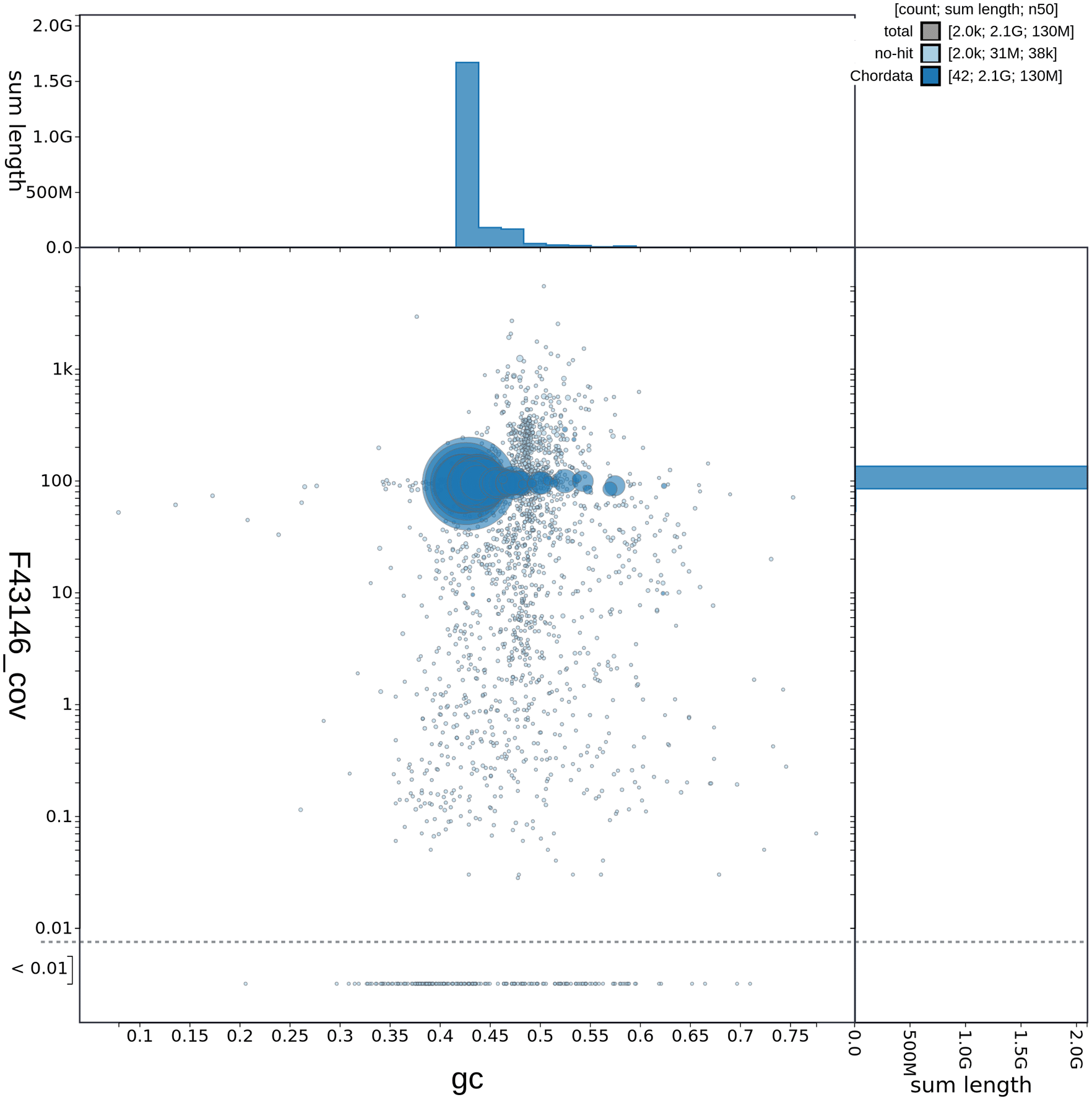
GC-coverage plot of C. caretta (rCarCar2) generated from Blobtoolkit v.2.6.4 (Challis et al., 2020). Scaffolds are coloured by phylum with Chordata represented by blue and no-hit represented by pale blue. Circles are sized in proportion to scaffold length. Histograms show the distribution of scaffold length sum along each axis.
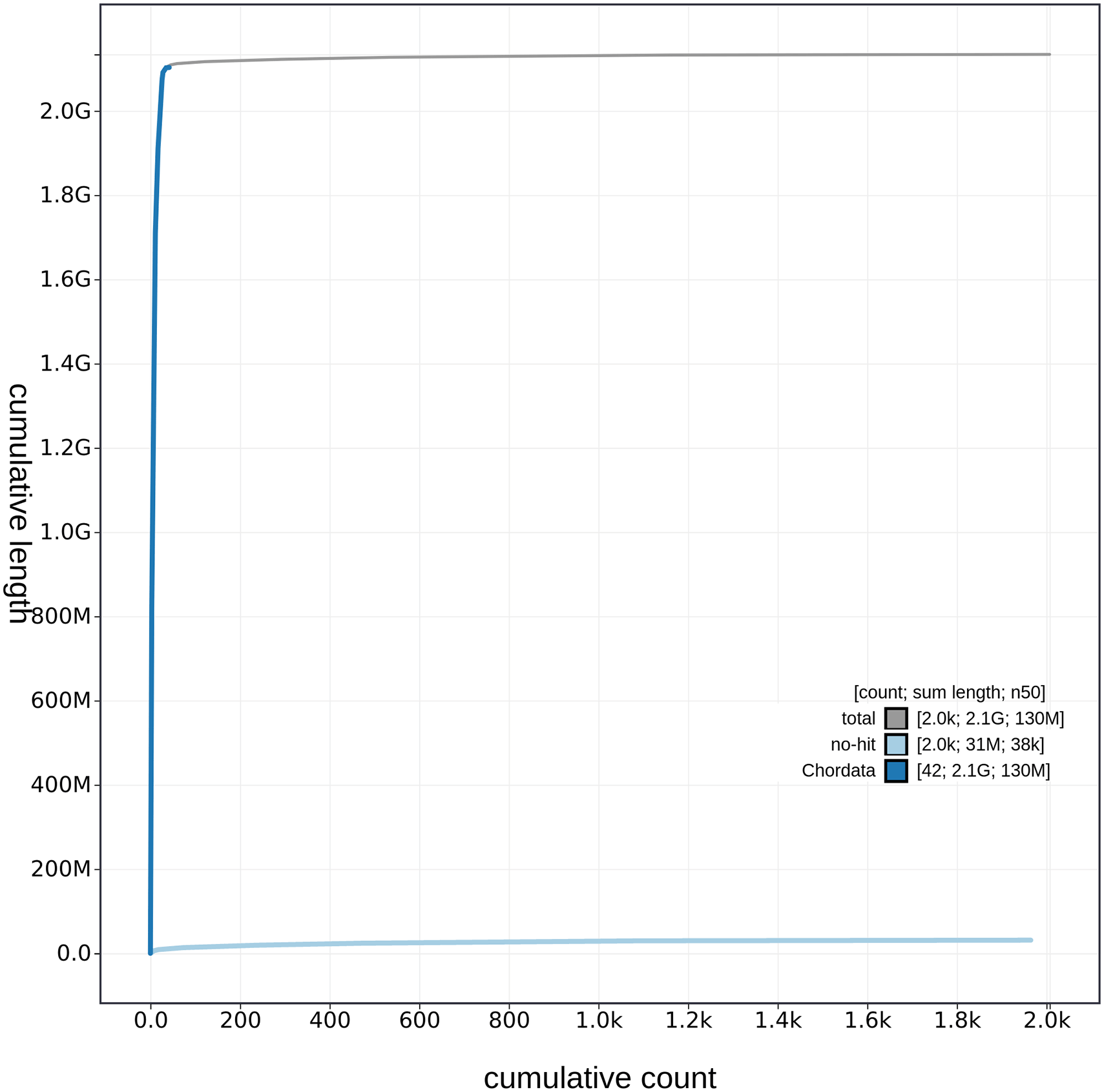
Cumulative sequence length of C. caretta (rCarCar2) generated from Blobtoolkit v.2.6.4 (Challis et al., 2020). The grey line shows the cumulative length for all scaffolds. Coloured lines show cumulative lengths of scaffolds assigned to each phylum using the BUSCO genes tax rule, with Chordata represented by blue and no-hit represented by pale blue.
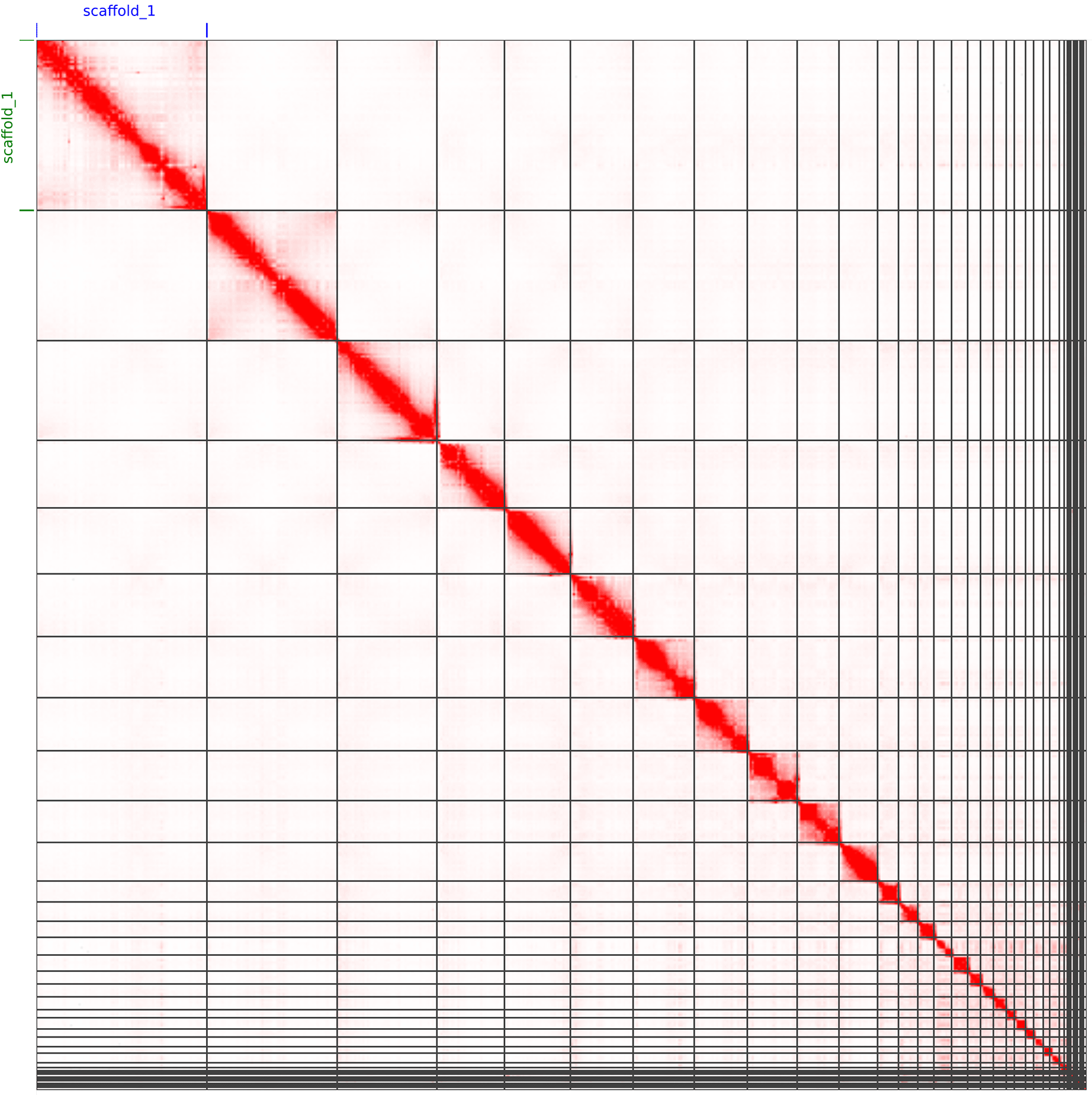
HiC contact map of rCarCar2 assembly visualized using JuiceBox v2.13.07 (Durand et al., 2016a). Chromosomes are shown in order of size from left to right and top to bottom. As an additional confirmation for the quality of the assembly, the microchromosomes are visible as a cluster of spatially-associated contigs in the lower right, as reported in by Waters et al., 2021.
| Software | Version | Source |
|---|---|---|
| Racon | 1.4.13 | Vaser et al., 2017 |
| Medaka | 1.2.0 | https://github.com/nanoporetech/medaka |
| Pilon | 1.23 | Walker et al., 2014 |
| Salsa | 2.3 | Ghurye et al., 2019 |
| BlobToolKit | 2.6.4 (BTK pipeline) 3.1.0 (Blobtoolkit) | Challis et al., 2020 |
| nf-core/hic | 1.1.0 | Servant and Peltzer, 2019 |
| Juicer Tools | 2.13.06 | Durand et al., 2016b |
| Juice Box | 2.13.06 | Durand et al., 2016a |
| Redbean | 2.5 | Ruan and Li, 2019 |
| LongStitch | 1.0.1 | Coombe et al., 2021 |
| Jupiter Plot | 1.0 | Chu, 2021 |
| Busco | 5.2.2 | Manni et al., 2021 |
| Quast | 5.0.2 | Gurevich et al., 2013 |
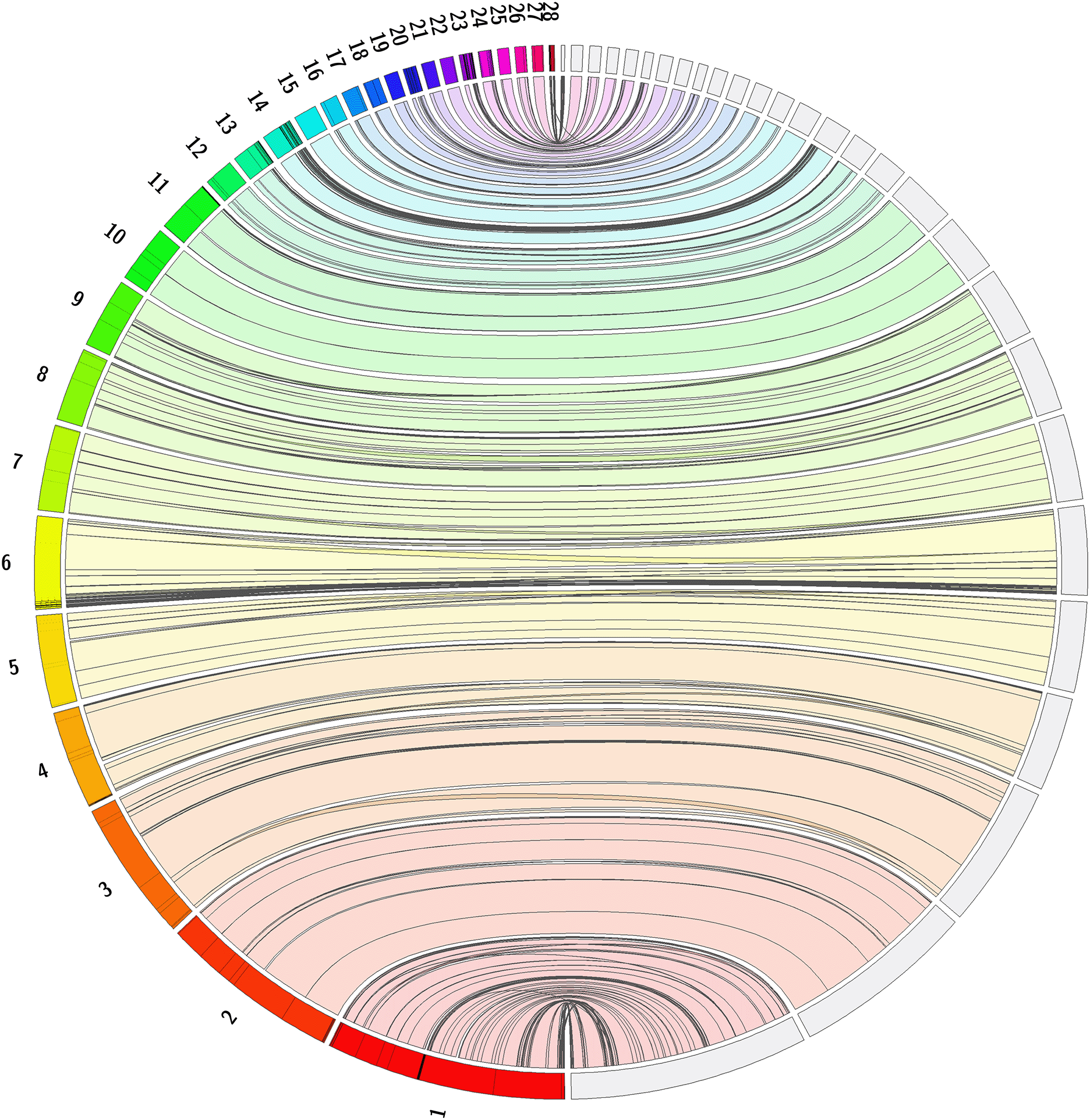
Full genome alignment of Caretta caretta genome, rCarCar2 (right), and Chelonia mydas (green sea turtle) genome (primary haplotype v2), rCheMyd1 (left), generated using Jupiter Plot (Chu, 2018). The left of the circle shows 28 green sea turtle chromosomes and the right of the circle shows 28 loggerhead sea turtle chromosomes. Coloured bands represent synteny between the genomes, and lines crossing the circle indicate genomic rearrangements, or break points in the scaffolds.
Annotation for the loggerhead sea turtle genome assembly (GSC_CCare_1.0 (GCA_023653815.1)) was generated by the Ensembl Rapid Release gene annotation pipeline (Aken et al., 2016). The resulting Ensembl annotation includes 42,302 transcripts assigned to 19,633 coding and 4,161 non-coding genes (Caretta caretta - Ensembl Rapid Release). The loggerhead sea turtle assembly was also annotated for 54,583 protein sequences using RefSeq (GCF_023653815.1, PRJNA853764).
National Centre for Biotechnology Information BioProject: Loggerhead Sea turtle (Caretta caretta) genome sequencing and assembly, rCarCar2. Accession number: PRJNA826225.
The genome sequence is released openly for reuse. The C. caretta genome sequencing initiative is part of the Canadian BioGenome Project and CanSeq150 Projects initiatives. All raw sequence data and the assembly have been deposited in INSDC databases. The genome is annotated through the Reference Sequence (RefSeq) database in BioProject accession number PRJNA853764. Raw data and assembly accession identifiers are reported in Table 1.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
No
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genomics, bioinformatics
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genomics, Bioinformatics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 27 Jun 23 |
read | |
|
Version 1 27 Mar 23 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)