Keywords
Plastid genome, Passifloraceae, Passiflora tripartita var. mollissima, poro-poro, native fruit, Huánuco, Peru
This article is included in the Genomics and Genetics gateway.
Plastid genome, Passifloraceae, Passiflora tripartita var. mollissima, poro-poro, native fruit, Huánuco, Peru
Passiflora tripartita var. mollissima (Kunth) Holms-Niels. & P.M. Jørg (ITIS, 2022) previously known as Passiflora mollissima (Kunth) Bailey (Primot et al., 2005), is a semi-perennial fruit plant (Mayorga et al., 2020). It is a diploid species with a small number of chromosomes (2n = 18) (Coppens D’Eeckenbrugge, 2001), which is placed in the section Elkea of supersection Tacsonia of subgenus Passiflora belonging to the Passifloraceae family (Segura et al., 2005; Ocampo & Coppens d’Eeckenbrugge, 2017). Poro-poro is a native fruit of the Andean region (Ocampo & Coppens d’Eeckenbrugge, 2017). It grows in the Peruvian highlands in the departments of Ancash, Junín, Moquegua, Huancavelica, and Huánuco at altitudes of 1,000–4,000 m.a.s.l. (Tapia & Fries, 2007; Ríos-García, 2017). It is widely used in traditional medicine (Ríos-García, 2017) and is considered one of the best Passiflora species based on its organoleptic characteristics (Primot et al., 2005). This fruit provides a source of vitamins (A, B3, and C) and minerals (magnesium, potassium, phosphorus, sodium, chlorine, iron, calcium, sulfur, zinc, copper, selenium, cobalt, and nickel) (Leterme et al., 2006; Chaparro-Rojas et al., 2014). In addition, it has an elevated antioxidant activity and high content of carotenoids (118.8 mg β-carotene), phenols (460.1 mg gallic acid), and flavonoids (1907.6 mg catechin/100 g) (Leterme et al., 2006; Chaparro-Rojas et al., 2014). Specifically, the high concentration of flavan-3-ols (a group of bioactive compounds) has been associated with beneficial effects on human health, such as cardiovascular protection, neurodegenerative diseases, and as an anti-cancer, anti-microbial, and anti-parasitic agent (Giambanelli et al., 2020; Luo et al., 2022).
Plastome sequences of more than 800 sequenced genomes are small in size with high copy numbers and conserved sequences, enabling a significant understanding of plant molecular evolution, structural variations, and evolutionary relationships of plant diversity (Daniell et al., 2016; Dobrogojski et al., 2020). The plastid genome has a quadripartite structure: a large single-copy (LSC) of 80–90 kilobase pairs (kb), a small single-copy (SSC) of 16–27 kb, and two sets of inverted repeats (IRa and IRb) of 20–28 kb, with 110–130 unique genes, including protein-coding genes, transfer RNA (tRNA), and ribosomal RNA (rRNA) (Ozeki et al., 1989; Wang & Lanfear, 2019). In recent years, declining genome sequencing costs resulted in more than 780 complete plant genomes of different species becoming available (Marks et al., 2021; Sun et al., 2022). Recently, some Passiflora plastid genomes such as Passiflora edulis (Cauz-Santos et al., 2017), Passiflora xishuangbannaensis (Hao & Wu, 2021), Passiflora caerulea (Niu et al., 2021), Passiflora serrulata (Mou et al., 2021), Passiflora foetida (Hopley et al., 2021), and Passiflora arbelaezii (Shrestha et al., 2019), became publicly available. However, despite the scarcity of genomic information on underutilized crops (Gioppato et al., 2019), we have only begun to investigate the genomics of plants of great importance for plant breeding programs. The aim of the present study was to sequence, assemble, and annotate the plastid genome of poro-poro to contribute to plant breeding programs. In the present study, we report the first plastid genome sequence submitted for an isolate of Passiflora tripartita var. mollissima from Peru, a species with great agro-industrial and pharmaceutical potential because of its beneficial characteristics for human health.
In November 2022, the fresh leaves of Passiflora tripartita var. mollissima were collected from Raccha Cedrón locality of Quisqui District, Huánuco Province from Peru (9°53′37″S, 76°26′02″W, altitude 2,945 m.a.s.l.). A herbarium voucher specimen (USM<PER>:MHN331530) was deposited in the Herbario San Marcos (USM) of the Museo de Historia Natural (MHN) at the Universidad Nacional Mayor de San Marcos (UNMSM) (see the Extended data, Aliaga et al., 2023a).
Total genomic DNA was extracted from approximately 100 mg fresh leaves (from voucher number USM<PER>:MHN331530) according to Doyle’s (1991) method with slight modifications. The DNA isolation buffer consisted of buffer cetyl-trimethyl ammonium bromide (CTAB) 3% (30g/L CTAB, 100 mM Tris-HCl pH 8.0, 10nM EDTA, 1.4 M NaCl, 0,2% 2-mercaptoethanol), 70% ethanol, chloroform-isoamyl alcohol (24:1), 10 mM ammonium acetate, isopropanol, TE buffer (10 mM Tris-H, 1 mM EDTA), and RNAase A (10 ug/ml). Genomic DNA quality was assessed using a fluorometry-based Qubit (Thermo Fisher Scientific, USA, catalog number: Q33238) coupled to a Broad Range Assay kit (Thermo Fisher Scientific, USA, catalog number: Q33230). High-quality DNA (230/260 and 260/280 ratios >1.8) were normalized (20 ng/μL) to examine its integrity using 1% (w/v) agarose gel electrophoresis (see the Extended data, Aliaga et al., 2023b) with the following equipment: Horizontal gel system (Fisher Scientific, Denmark, catalog number: 11833293, 150mm (length), 100 mm (width)), Transilluminator (Fisher Scientific, Spain, catalog number: 12864008), and digital camera (Canon, Spain, catalog number: 2955C002); Reagents: TAE buffer (40 mM Tris, 20mM NaAc, 1mM EDTA, pH 7.2), loading buffer 6X (Promega, USA, catalog number: G1881, 0.4% orange G, 0.03% bromophenol blue, 0.03% xylene cyanol FF, 15% Ficoll® 400, 10mM Tris-HCl pH 7.5 and 50mM EDTA pH 8.0) and Ethidium bromide (Promega, USA, catalog number H5041, 10 mg/ml), and 1 Kb Plus DNA Ladder (ThermoFisher, USA, catalog number: 10787018).
Qualified DNA was fragmented, and the TruSeq Nano DNA kit (Illumina, San Diego, CA, USA, catalog number: FC-121-4001) was used to construct an Illumina paired-end (PE) library. PE sequencing (2 × 150 bp) was performed using the Illumina NovaSeq 6000 platform (Modi et al., 2021) (Illumina, San Diego, Ca, USA, catalog number: 20012850) (Macrogen, Inc., Seoul, Republic of Korea). All adapters and low-quality reads were removed using the FastQC (Wingett & Andrews, 2018) and Cutadapt (Martin, 2011) programs. PE reads (2 × 150 bp) were evaluated for quality using QUAST (Gurevich et al., 2013) analysis, and subsequent steps used clean data. Then, clean reads obtained were assembled into a circular contig using NOVOPlasty v.4.3 (Dierckxsens et al., 2017), with P. edulis (NC_034285) as the reference (Cauz-Santos et al., 2017). The plastid genome was annotated using the Dual Organellar GenoMe Annotator GeSeq (Tillich et al., 2017) and CpGAVAS2 (Shi et al., 2019). A circular genome map was constructed using OGDRAW v.1.3.1 (Greiner et al., 2019). Finally, the completed sequences were submitted to the NCBI GenBank under the accession number OQ910395 (GenBank, 2023).
We used 26 complete plastome sequences to infer the phylogenetic relationships among Passiflora species, and Vitis vinifera was used as an outgroup (see the Extended data, Aliaga et al., 2023c). Single-copy orthologous genes were identified using the Orthofinder version 2.2.6 pipeline (Emms & Kelly, 2019). For each gene family, the nucleotide sequences were aligned using the L-INS-i algorithm in MAFFT v7.453 (Katoh & Standley, 2013). A phylogenetic tree based on maximum likelihood (ML) was constructed using RAxML v8.2.12 (Stamatakis, 2014) with the GTRCAT model. A phylogenetic ML tree was reconstructed and edited using MEGA 11 (Tamura et al., 2021) with 1000 replicates.
The plastid genome sequences of P. tripartita var. mollissima (poro-poro) (Figure 1) was 163,451 bp in length, with a typical quadripartite structure consisting of a large single-copy (LSC) region of 85,525 bp (52.32% in total) and a small single-copy (SSC) region of 13,518 bp (8.27%), separated by a pair of inverted repeat regions (IRs) of 32,204 bp (19.70%). The poro-poro plastome is 12,045 bp longer than that of one of the most economically important species, passion fruit (P. edulis) (Cauz-Santos et al., 2017), and is only 7,117 bp longer than that of the longest Passiflora plastome reported, i.e., P. arbelaezii (Shrestha et al., 2019). The plastome sequence of poro-poro has a similar quadripartite architecture to other plants (Ohyama et al., 1986; Shinozaki et al., 1986; Nguyen et al., 2021). However, the LSC region is 4,150 bp longer than that of P. xishuangbannaensis but is 98bp, 195 bp, and 1,927 bp shorter than that of P. caerulea, P. edulis, and P. arbelaezii, respectivety. The SSC region is 121 bp, 140 bp, 359 bp, and 754 bp longer than that of P. caerulea, P. edulis, P. xishuangbannaensis, and P. arbelaezii, respectively. The IRs regions are 6,024 bp, 6,050 bp, and 11,600 longer than that of P. caerulea, P. edulis, and P. xishuangbannaensis, respectively; however, it is 2,972 bp shorter than that of P. arbelaezii (Cauz-Santos et al., 2017; Shrestha et al., 2019; Hao & Wu, 2021; Niu et al., 2021). The plastome structure of the P. tripartita var. mollissima consisted of A = 30.79%, T(U) = 32.34%, C = 18.67% and G = 18.20%. The overall AT content of the plastid genome was 63.13%, whereas the overall GC content was 36.87% as similar to that of other reported chloroplast genomes from the same family, such as 36.90% in P. arbelaezii (Shrestha et al., 2019), 37% in P. edulis and P. serrulata (Cauz-Santos et al., 2017; Mou et al., 2021), 37.03% in P. caerulea (Niu et al., 2021), and 37.1% in P. xishuangbannaensis (Hao & Wu, 2021).
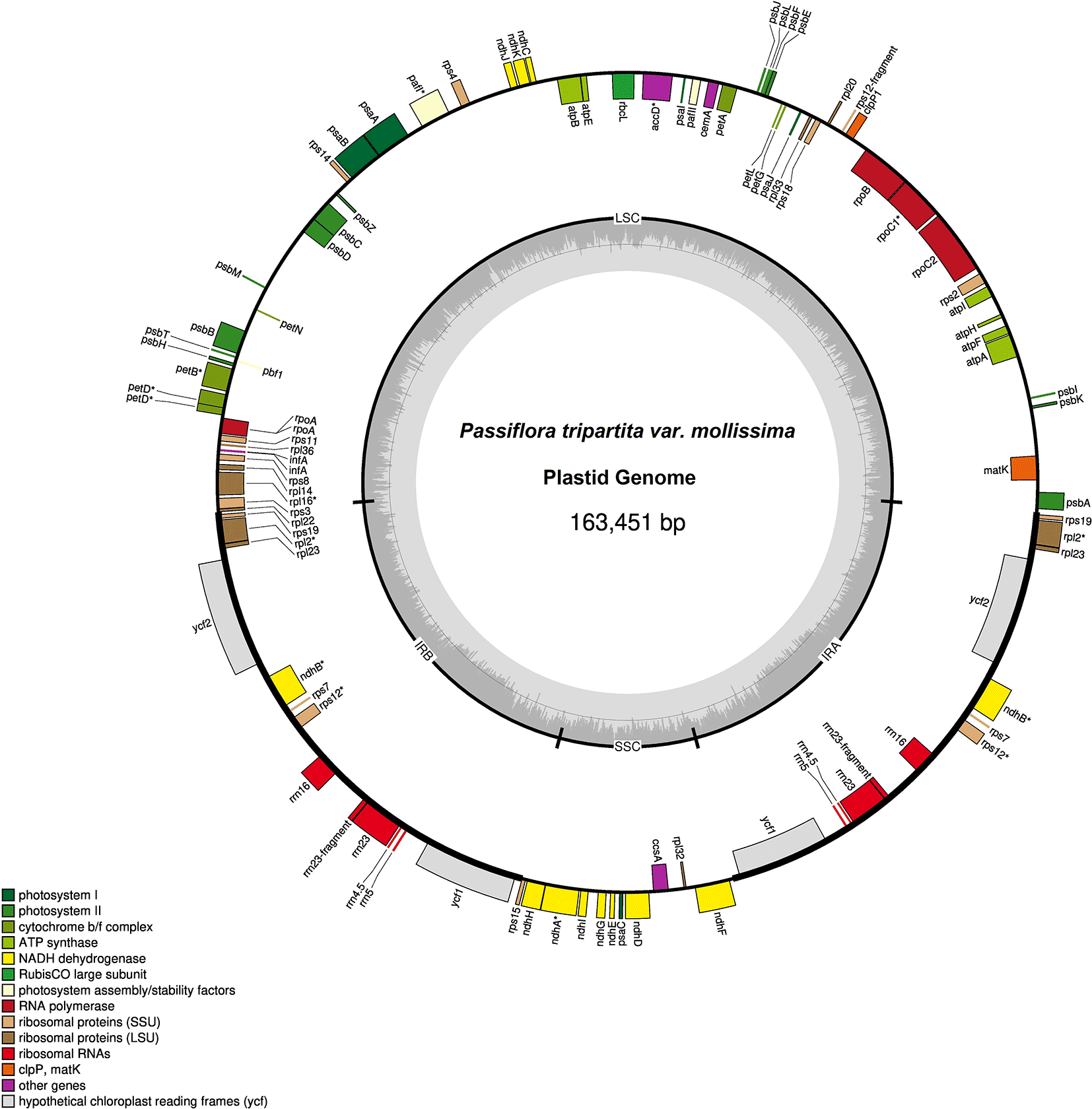
The thick lines indicate the IR1 and IR2 regions, which separate the large single-copy (LSC) and small single-copy (SSC) regions. Genes marked inside the circle are transcribed clockwise, and genes marked outside the circle are transcribed counterclockwise. Genes are color-coded based on their function, shown at the bottom left. The inner circle indicates the inverted boundaries and guanine and cytosine (GC) content.
Poro-poro plastid genome annotation identified 129 genes, of which 112 were unique, and 17 were duplicated in the inverted repeat (IR) region. The plastome contained 85 protein-coding genes, 37 transfer RNA (tRNA)-coding genes, seven ribosomal RNA (rRNA)-coding genes, and 14 genes with introns (12 genes with one intron and two genes with two introns), as shown in Table 1. The poro-poro plastid genome contained 112 unique genes, of which there were 29 tRNA genes, four rRNA genes, and 79 protein-coding genes. The latter comprised 20 ribosomal subunit genes (nine large subunits and 11 small subunit), four DNA-directed RNA polymerase genes, 46 genes were involved in photosynthesis (11 encoded subunits of the NADH oxidoreductase, seven for photosystem I, 15 for photosystem II, six for the cytochrome b6/f complex, six for different subunits of ATP synthase, and one for the large chain of ribulose biphosphate carboxylase), eight genes were involved in different functions, and one gene was of unknown function (Table 2).
| Features | Poro-poro1 |
|---|---|
| Genome size (bp) | 163,451 |
| aLSC length (bp) | 85,525 |
| bSSC length (bp) | 13,518 |
| cIR length (bp) | 32,204 |
| Total GC content (%) | 36.87 |
| dA content (%) | 30.79 |
| eT(U) content (%) | 32.34 |
| fG content (%) | 18.20 |
| gC content (%) | 18.67 |
| Total number of genes | 129 |
| Protein-coding genes | 85 |
| hrRNA coding genes | 7 |
| itRNA coding genes | 37 |
| Genes duplicated in IR regions | 17 |
| Total introns | 14 |
| Single introns (gene) | 12 |
| Double introns (gene) | 2 |
| Group of genes | Gene names |
|---|---|
| Photosystem I | psaA, psaB, psaC, psaI, psaJ, ycf3 **, ycf4 |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| Cytochrome b/f complex | petA, petB, petD *, petG, petL, petN |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI |
| NADH dehydrogenase | ndhA*, ndhB * (X2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| RubisCO large subunit | rbcL |
| DNA-dependent RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 |
| Ribosomal proteins (SSU) | rps2, rps3, rps4, rps8, rps11, rps12 ** (X2), rps14, rps15, rps16, rps18, rps19 (X2) |
| Ribosomal proteins (LSU) | rpl2 * (X2), rpl14, rpl16 *, rpl20, rpl22, rpl23 (X2), rpl32, rpl33, rpl36 |
| Acetyl-CoA carboxylase | accD |
| C-type cytochrome synthesis | ccsA |
| Envelope membrane protein | cemA |
| Protease | clpP |
| Translational initiation factor IF-1 | infA |
| Maturase | matK |
| Component of TIC complex | yct1, ycf2 |
| Unknown function protein-coding | ycf15 (X2) |
| Ribosomal RNAs | rrn4.5, rrn5 (X2), rrn16 (X2), rrn23 (X2) |
| Transfer RNAs | trnA-UGC * (X2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC *, trnH-GUG, trnI-CAU (X2), trnI-GAU * (X2), trnK-UUU *, trnL-CAA (X2), trnL-UAA *, trnL-UAG, trnM-CAU (X2), trnN-GUU (X2), trnP-UGG, trnQ-UUG, trnR-ACG (X2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC (X2), trnV-UAC *, trnW-CCA, trnY-GUA |
In the plastid genome, 14 genes contained introns distributed as follows: the LSC, SSC, and IRs regions contained eight genes (petD, rpl16, rpoC1, trnG-UCC, trnK-UUU, trnL-UAA, trnV-UAC, and ycf3), one gene (ndhA), and five genes (ndhB, rpl2, rps12, trnA-UGC, and trnI-GAU) respectively. Similarly, these genes included six protein-coding genes, each with a single intron (petD, ndhA, ndhB, rpoC1, rpl2, and rpl16); six tRNA genes, each with a single intron (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC); and two protein-coding genes with two introns (ycf3 and rps12). Except for 17 genes that were duplicated in the IR region (ndhB, rps19, rpl2, rpl23, rps12, ycf15, rrn5, rrn16, rrn23, trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnM-CAU, trnN-GUU, trnR-ACG, and trnV-GAC) all genes contained a single copy, as shown in Table 2. The plastome of P. tripartita var. mollissima contained eight genes (ycf1, ycf2, ycf15, rps16, rpl20, rpl22, accD, infA) that were lost or non-functional genes in P. edulis; and compared to P. edulis, it has one absent gene (trnfM-CAU), as previously reported (Cauz-Santos et al., 2017). In this study, the ycf1 sequence encodes a protein essential for plant viability and a vital component of the translocon on the inner chloroplast membrane (TIC) complex (Kikuchi et al., 2013), and ycf2 is a component of the ATPase motor protein associated with the TIC complex (Kikuchi et al., 2018).
To identify the evolutionary position of Passiflora tripartita var. mollissima in the Passifloraceae family, phylogenetic relationships based on the OrthoFinder clustering method were used to avoid erroneous rearrangements in phylogenetic tree reconstruction and provides a more reliable evolutionary analysis (Gabaldón, 2005; Zhang et al., 2012). The phylogenetic tree was constructed based on single-copy orthologous genes (Emms & Kelly, 2019) and maximum likelihood analysis with the complete annotated protein sequences of 27 plastid genomes, of which 26 were from Passiflora species. One species, Vitis vinifera, was chosen as the outgroup.
Maximum likelihood (ML) bootstrap values ranged from 38%–92% for seven of the 25 nodes. All nodes except the indicated ones (seven nodes) exhibited bootstrap support (BS) values of 100%. These Passiflora species were divided into four groups: subgenus Passiflora (P. nitida, P. quadrangularis, P. cincinnata, P. caerulea, P. edulis, P. laurifolia, P. vitifolia, P. serratifolia, P. serrulata, P. ligularis, P. serratodigitata, P. actinia, P. menispermifolia and P. oerstedii), subgenus Tetrapathea (P. tetrandra), subgenus Decaloba (P. microstipula, P. xishuangbannaensis, P. biflora, P. lutea, P. jatunsachensis, P. suberosa and P. tenuiloba), and subgenus Deidamoides (P. contracta and P. arbelaezii). The relationships between the four subgenera of Passiflora species (Passiflora, Tetrapathea, Decaloba, and Deidamoides) were congruent and strongly supported by the same patterns as previously reported (Cauz-Santos et al., 2020; Pacheco et al., 2020). These results resolved Passiflora tripartita var. mollissima belonging to the subgenus Passiflora, which was closely related to P. menispermifolia and P. oerstedii with 100% BS, and was sister to P. tetrandra (subgenus Tetrapathea), P. biflora (subgenus Decaloba), and P. contracta (subgenus Deidamoides), as shown in the cladogram (Figure 2).
Nucleotide: Passiflora tripartita var. mollissima chloroplast, complete genome. Accession number: OQ910395. https://identifiers.org/nucleotide:OQ910395 (GenBank, 2023).
Figshare: Herbarium specimen voucher of Passiflora tripartita var. mollissima (Kunth) Holms-Niels. & P.M. Jørg (USM:MHN331530). https://doi.org/10.6084/m9.figshare.23556654 (Aliaga et al., 2023a).
Figshare: Gel imagen of DNA isolate from poro-poro sample. https://doi.org/10.6084/m9.figshare.23560755 (Aliaga et al., 2023b).
Figshare: Details of the plastid genome sequences used for phylogenetic analysis. https://doi.org/10.6084/m9.figshare.23556834 (Aliaga et al., 2023c).
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
We thank the Universidad Privada del Norte (UPN) for funding the APC. We thank the Servicio Nacional Forestal y de Fauna Silvestre (SERFOR) for authorized this research project (AUT-IFL-2022-058). We thank Prof. Dr. Esteban Hopp (Universidad de Buenos Aires) for their careful reading of the manuscript and their constructive remaks. We thank Petr Sklenář and Filip Kolar for their help in the sample collection. We thank curator Julio C. Torres–Martinez (Museo de Historia Natural, Universidad Nacional Mayor de San Marcos) for the taxonomy identification and deposit of the plant specimen. We thank Dr. Rajest Mahato and Dr. Giusseppe D’Auria for the recommendations and bioinformatics support. We thank Mr. Julián Vasquez-Arriaga for administrative support (Plant Science Laboratory).
An earlier version of this article can be found on Preprints.org (doi: https://doi.org/10.20944/preprints202306.0463.v2).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Are the rationale for sequencing the genome and the species significance clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
Partly
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: I can assess all aspects of the manuscript and published up to 18 articles in the same field.
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Organelle genome sequencing, Transcriptome assembling, Genetic diversity, Barcoding and DNA markers etc
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 3 (revision) 19 Feb 24 |
read | |
|
Version 2 (revision) 09 Jan 24 |
read | read |
|
Version 1 07 Jul 23 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)