Keywords
Biocontrol agent; phylogenetic analysis; Rhizospheric region; Species diversity; Nepal
This article is included in the Agriculture, Food and Nutrition gateway.
Trichoderma spp. hold significant potential as biocontrol agents in agriculture due to their antagonistic properties against plant pathogens. The study aimed to characterize and identify Trichoderma isolates from rhizospheric regions of vegetable crops.
In this study, Trichoderma isolates were collected from rhizospheric soil samples of vegetable crops from different ecological zones and were selected for comprehensive morphological and molecular characterization. The isolates were visually assessed for colony color, growth pattern, aerial mycelium presence, phialide and conidial morphology, and chlamydospore presence. Molecular analysis was employed based on ITS and tef-1α sequences. Diversity indices were also computed for different ecological zones.
The morphological characteristics and phylogenetic trees for both regions provided a clear species resolution, with four main clades: Harzianum, Viride, Brevicompactum and Longibrachiatum with 12 species T. harzinaum, T. afroharzianum, T. lentiforme, T. inhamatum, T. camerunense, T. azevedoi, T. atroviride, T. asperellum, T. asperelloides, T. koningii, T. longibrachiatum and T. brevicompactum and nine species as a new country record. Diversity indices indicated that high mountain regions displayed the highest species diversity and evenness (H = 1.724 [0.28], J = 0.84, D = 0.28), followed by hilly regions (H = 1.563 [0.28], J = 0.72, D = 0.28). Plains, on the other hand, exhibited lower species diversity (H = 1.515, J = 0.66, D = 0.33). The calculated species abundance values showed that plains (E = 2.11), mid-hills (E = 1.95), and high mountains (E = 1.99) each had their unique diversity profiles. Notably, T. afroharzianum and T. asperellum were predominant.
Overall, the study unveiled a rich diversity of Trichoderma species in different agricultural zones of Nepal. These findings shed light on the ecological distribution and diversity of Trichoderma spp., which could have significant implications for sustainable agriculture and biological control strategies.
Biocontrol agent; phylogenetic analysis; Rhizospheric region; Species diversity; Nepal
The genus Trichoderma comprises a highly diverse group of soil dwelling fungi, demonstrating remarkable versatility. Within this genus, a wide array of strains exhibits significant potential for various applications, including biological control as biopesticides, biofertilizers, soil enhancers and promoters of crop growth.1,2 Furthermore, Trichoderma isolates have the capacity to stimulate plant defense mechanisms, a phenomenon extensively documented in studies conducted by various researchers.3–6
The Trichoderma genus was originally proposed by Persoon in Germany in 1794, more than two centuries ago, and their potential as bioagents was first documented by Weindling in 1932.7 To date, over 250 Trichoderma species have been identified worldwide.8 Trichoderma species can be readily isolated from soil using various conventional methods, thanks to their adaptability to diverse conditions, ranging from deserts and wetlands to the Himalayan regions. However, the most effective method for isolating Trichoderma spp. is from rhizospheric soil samples.9 A comprehensive understanding of Trichoderma biodiversity is crucial for harnessing the full potential of this genus in various biological applications. Previous surveys that investigated Trichoderma diversity in specific geographical regions have primarily focused on isolates obtained from soil samples. These surveys have been conducted in various locations, including Russia and the Siberian Himalayas,10,11 China,12,13 South-East Asia,14 India,15 Egypt,16 Iran,17 Philippines,18 Europe,19 Canary Islands,20 Sardinia21and South America.22 The potential of Trichoderma species as biological control agents (BCAs) has been under investigation for over 70 years, and more recently, these species have become commercially available.23,24 Certain Trichoderma species have been successfully utilized as BCAs, showcasing their antagonistic mechanisms to mitigate damage caused by soil-borne pathogens, both in greenhouse and open field conditions.25–27 Trichoderma deploys diverse mechanisms, including antibiosis, competition, mycoparasitism, and enzymatic hydrolysis, to manifest its biocontrol activity.28 However, it is important to note that not all Trichoderma species yield the same effects on crops or pathogens. Therefore, the characterization and precise identification of isolates at the species level are pivotal for their effective application as BCAs and the preservation of isolates as viable commercial products.24
Traditional methods for identifying Trichoderma species have conventionally relied on morphological characteristics and culture-based approaches. However, owing to the limited and often indistinguishable morphological features shared among Trichoderma species, alongside the increasing recognition of morphologically cryptic species, accurate identification through these methods becomes challenging and can potentially lead to misidentification.10,29 As a result, molecular techniques such as amplification of important taxonomic markers, and sequencing have emerged as indispensable tools for achieving precise species identification.29–32 In the present study, the identification of Trichoderma species at the species level was accomplished by focusing on two key genetic markers: the Internal Transcribed Spacer (ITS) region of rDNA and a partial sequence of the translation elongation factor 1-alpha gene (tef-1α) in addition to classical taxonomy.33 Recent researches have evaluated the ecological specialization of diversity. Numerous studies have documented the discovery of new isolates and phylogenetic species across various natural ecosystems.20,34 While some studies have been conducted in Nepal, they all have focused on different ecological zones.35 In contrast, limited attention has been given to studying Trichoderma in agricultural environments. Importantly, such investigations have practical implications, as they highlight the potential of agricultural soil rhizosphere as a rich source of beneficial strains with biocontrol capabilities. Building upon these findings, we propose that the composition, distribution, and genetic structure of Trichoderma spp. in the rhizosphere of vegetable crops along with ecological domains and altitudinal gradients may exhibit significant differences. Confirming these differences will enhance our understanding of Trichoderma’s biodiversity across distinct biological niches and various altitudinal gradients across the country.
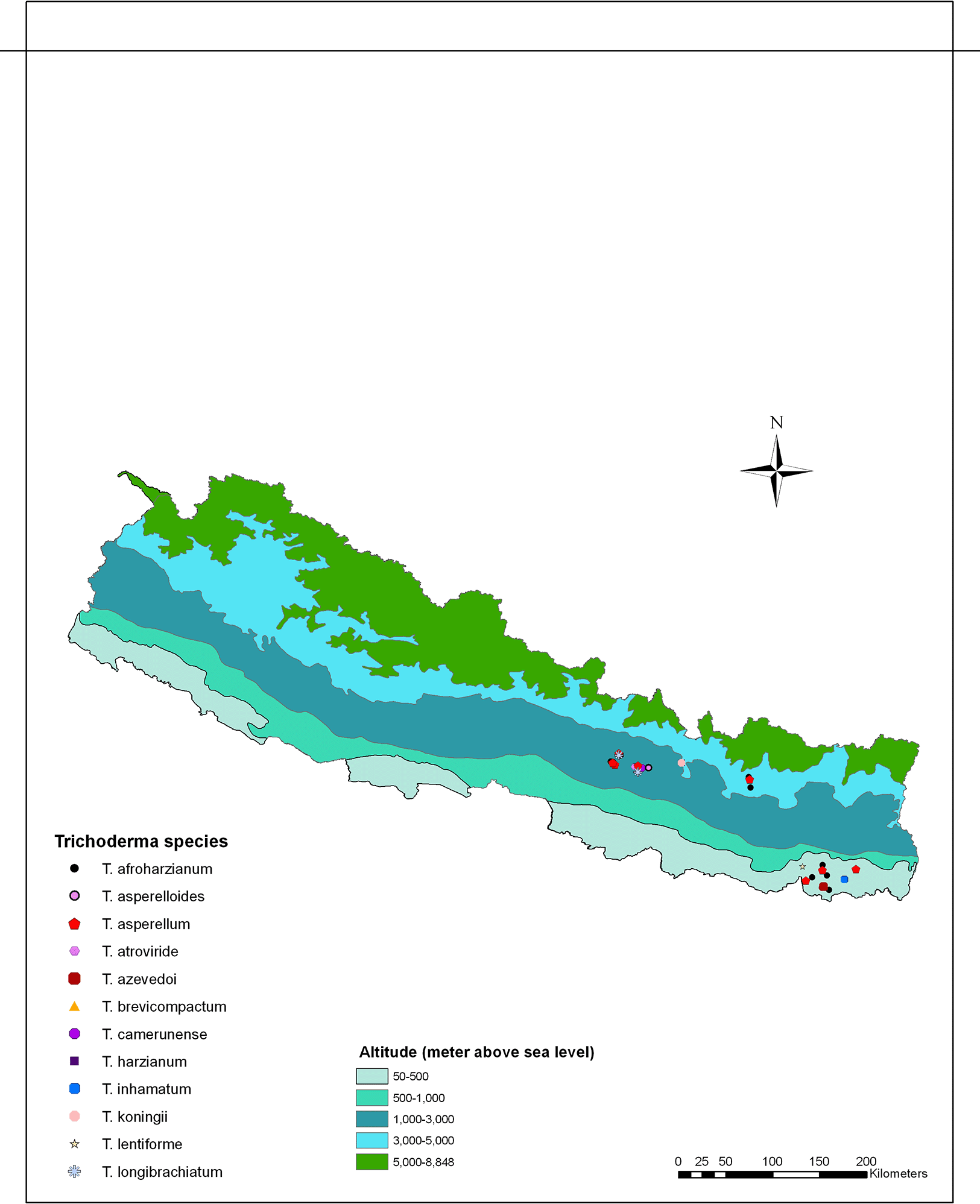
To isolate Trichoderma, soil samples were collected from different agricultural regions spanning three ecological zones in Nepal: the plain region (80-141 m above sea level), the mid-hill region (1284-1650 m above sea level), and the high mountain region (1800-2500 m above sea level). These regions exhibited variations in altitude and ecological traits, and were recognized for cultivating a range of agricultural crops. The soil samples were collected from vegetable-growing areas within each region, covering eight districts in Nepal, including Sindhupalchok and Solukhumbu districts in the high mountains; Kathmandu, Lalitpur, Bhaktapur and Kavrepalanchok districts in the mid-hills, and Sunsari and Morang in the plains (Figure 1).
Using a sterile spatula, approximately 200 grams of soil were collected from the rhizospheric zone of crops such as cauliflower, leaf mustard, cabbage, asparagus bean, tomato, cucumber, brinjal, chilly, pumpkin, and okra. These soil samples were carefully placed in clean polythene bags and appropriately labelled. Subsequently, the soil samples were transported to the National Plant Pathology Research Center, Khumaltar, Nepal and stored at 4°C until isolation.
For isolation, 10 grams soil samples were suspended in 90 ml of sterile distilled water and mixed using a rotary shaking machine set at 260 × g for 30 minutes to ensure thorough homogenization.36 To facilitate the isolation, 1 mL aliquots of the soil suspensions were diluted from 10−1 to 10−5 onto Trichoderma Selective Media (TSM) using the spread plate technique as described by Askew and Laing.37 After inoculation, Trichoderma colonies were sub-cultured onto potato dextrose agar (PDA) plates and incubated at 28±2°C for seven days to allow for development and growth. Colonies have key characteristics that can be used to identify them as Trichoderma including growth pattern, growth speed, odour and colour. Depending on the diversity of colony characteristics, one to two colonies were selected from each sample for pure culture preparation. Each individual isolate was assigned a unique code and stored in a PDA slant for further study. Isolates were then grown on PDA plates for 3-5 days for sporulation. Different dilutions (10−1 to 10−3) of spore suspension for each isolate were spread on 10% water agar plates and incubated at 28±2°C for 24 hours in the dark to prepare monoconidial cultures. A germinating, isolated single spore was picked up using a sterile needle from the water agar plates and placed on PDA plates to prepare monoconidial culture for each isolate. Spore suspensions were prepared as mentioned earlier and preserved in silica gel for long term storage at -40°C.38
A total of 167 Trichoderma isolates were isolated from various soil samples and preserved using silica gel and were subsequently cultured on PDA plates at 28±2°C for three days. Following the incubation period, cultures with similar morphological characteristics, including growth pattern, diffusible pigments and odor, were observed and sorted into 25 different groups (T1-T25). While most groups consisted of 4 to 5 isolates, T11 and T22 (two larger groups) comprised 20 to 25 isolates each. Within each group, one representative isolate was selected for further study, except for the larger groups where 3 to 5 isolates were chosen for the study. Morphological characteristics such as the aspects of phialides, conidia and chlamydospores were scrutinized under a light microscope (Optika Microscope Italy, B-383PLi) equipped with a camera. To achieve this, Trichoderma isolates were grown on corn meal agar plate for 2-3 days, after which mycelial tips from the colony margin were transferred onto microscope slide and mounted with 3% KOH using sterile glass needles. Subsequently, these specimens were subjected to observation following the flattening and stretching of hyphae with a pair of glass needles. Once suitably prepared, a coverslip was placed over the samples, and any excess liquid was removed using tissue paper. Microscopical measurements and analyses were conducted using the ImageJ software (https://imagej.en.download.it, LOCI, University of Wisconsin). A total of 30 individual phialides and conidia from each specimen were measured and analyzed.
The colony characteristics of Trichoderma isolates were investigated on PDA plates that were incubated at 28°C under dark conditions. Notably, all Trichoderma isolates developed conidia within 4-5 days. Various diffusible pigments were observed, manifesting as distinct colors, which were then compared with the Audrey & Bear Color Chart (https://www.audrey and bear.com). Additionally, the presence or absence of a coconut odor was noted following 48 hours of culturing on PDA plates.39
Genomic DNA was extracted from thirty-three isolates, which morphologically represented different Trichoderma species from various ecological regions (Figure 2). Approximately, 30-40 mg of mycelium was scraped off from 3–5-day-old colonies cultured on potato dextrose agar (PDA) using sterilized glass slides prior to sporulation. The obtained mycelia were then ground in liquid nitrogen and DNA extraction was carried out using the Promega Wizard genomic DNA extraction kit following the manufacturer’s instructions (Promega Corporation, Madison, WI).
A fragment of rRNA containing the internal transcribed spacer regions was amplified using the primers ITS4 (TCCTCCGCTTATTGATATGC) and ITS5 (GGAAGTAAAAGTCGTAACAAGG).40 The translation elongation factor 1-alpha gene (tef-1α) was amplified using the primers EF1-728F (CATCGAGAAGTTCGAGAAGG)41 and TEF1R (GCCATCCTTGGGAGATACCAGC).10,42,43 Each PCR reaction (25 μL) comprised 12.5 μL of GoTaq® Green Master Mix (Promega), 9.5 μL of nuclease-free water, 1 μL of each forward and reverse primer (10 mM), and 1 μL of gDNA. The PCR reactions were performed using a thermocycler (LifeEco Thermal Cycler (BIOER)) with the following settings: For ITS: 1 cycle of 2 minutes at 95°C followed by 34 cycles of 30 seconds at 95°C, 30 seconds at 57°C, 1 minute at 72°C, followed by a final elongation step at 72°C for 5 minutes. For tef-1α: 1 cycle of 5 minutes at 95°C followed by 35 cycles of 45 seconds at 95°C, 45 seconds at 63°C, 1 minute at 72°C, followed by a final elongation step at 72°C for 10 minutes. Amplified PCR products (3 μL) were subjected to electrophoresis (45 minutes, 100 volts) in a 1.0% agarose gel stained with GelGreen® Nucleic Acid Gel Stain (Biotium, Fremont, CA, USA) alongside a 1 kb DNA ladder (Promega) for size estimation of the amplified bands. The PCR products were purified using the Wizard SV gel and PCR cleanup system (Promega) following the manufacturer’s instructions and were subsequently sequenced using both reverse and forward primers by Sanger sequencing (BGI Solutions Co., Ltd. Tai Po, Hong Kong, China).
Sequences were manually edited using Finch TV 1.4.0 for Windows (Geospiza, https://digitalworldbiology.com/FinchTV). Consensus sequences of forward and reverse amplicons were created using BioEdit software44 and deposited in the NCBI under accession numbers (OR140790- OR140818) for ITS rRNA and (OR567133-OR567158) for tef-1α. The BLASTn similarity search program was employed to identify homologous sequences in the NCBI nucleotide database, confirming species-level similarity with the sequence of the isolates. The ITS and tef-1α gene sequences were concatenated using Mesquite software version 2.75.45 Final alignments were done using Clustal W 2.0 in MEGA X version 10.1 and were used to construct phylogenetic trees using the Maximum Likelihood method with the Tamura-Nei evolution model.46 The choice of the Tamura-Nei evolution model was made after evaluating individual alignments through a model test (MEGAX software). Bootstrap values were computed with 1,000 replications to assess the statistical support for each branch. Graphical representation and editing of the phylogenetic trees were carried out using the Interactive Tree of Life (v3.5.1).47,48
Analysis of the diversity among Trichoderma species
The degree of dominance index, also known as the McNaughton dominance index (Y), was used to quantitatively assess the habitat preference of Trichoderma isolates in different agricultural fields. The dominance values were calculated using the equation:
where ‘N’ is the total number of Trichoderma isolates, ‘ni’ is the number of the genus (species) i, and ‘fi’ is the frequency with which genus (species) i appears in the samples. A species i is considered dominant when Y > 0.02.49Species richness (the total number of species), abundance (the total number of isolates of each species), and diversity were calculated using several ecological indices: the Simpson biodiversity index (D),50 Shannon’s biodiversity index (H),51 Pielou species evenness index (J),52 and Margalef’s abundance index (E).53 These indices were applied to quantitatively describe the diversity and habitat preferences of Trichoderma species in various agricultural fields across different ecological zones of Nepal.
The biological diversity indices were calculated using the following equations:
where Ni = 1, and Pi =In these equations, ‘S’ is the total number of Trichoderma species, ‘N’ is the total number of Trichoderma species isolates, ‘Pi’ is the relative quantity of Trichoderma species ‘i’, and ‘ni’ is the number of isolates of Trichoderma species ‘i’.
A total of 167 Trichoderma isolates were isolated from soil samples collected from rhizospheric regions of various vegetable crops. Among this collection, 33 Trichoderma isolates were selected based on morphological and microscopic characteristics. The isolates were visually characterized based on phenotypic traits such as colony color (ranging from dark green to cottony whitish green, Plate 1), growth pattern, presence or absence of aerial mycelium (Figure 3a, b), as well as the shape and size of phialides (Figure 3c–f) and conidia (Figure 3g–i). Additionally, the presence (Figure 3j) or absence of chlamydospores was observed microscopically (Table 1). Isolates from the groups T2, T3, T6, T7, T8, T9, T11, T14, T18, T19, T24 were morphologically and microscopically identified as belonging to the Harzianum clade. Meanwhile, isolates from groups T1, T4, T10, T12, T13, T15, T16, T21, T22, T23, T25 were identified as members of the Viride clade. Isolates from groups T5 and T17 were grouped within Longibrachiatum clade, corresponding to T. longibrachiatum species, while isolates from T20 group were classified within Brevicompactum clade, associated with T. brevicompactum species.
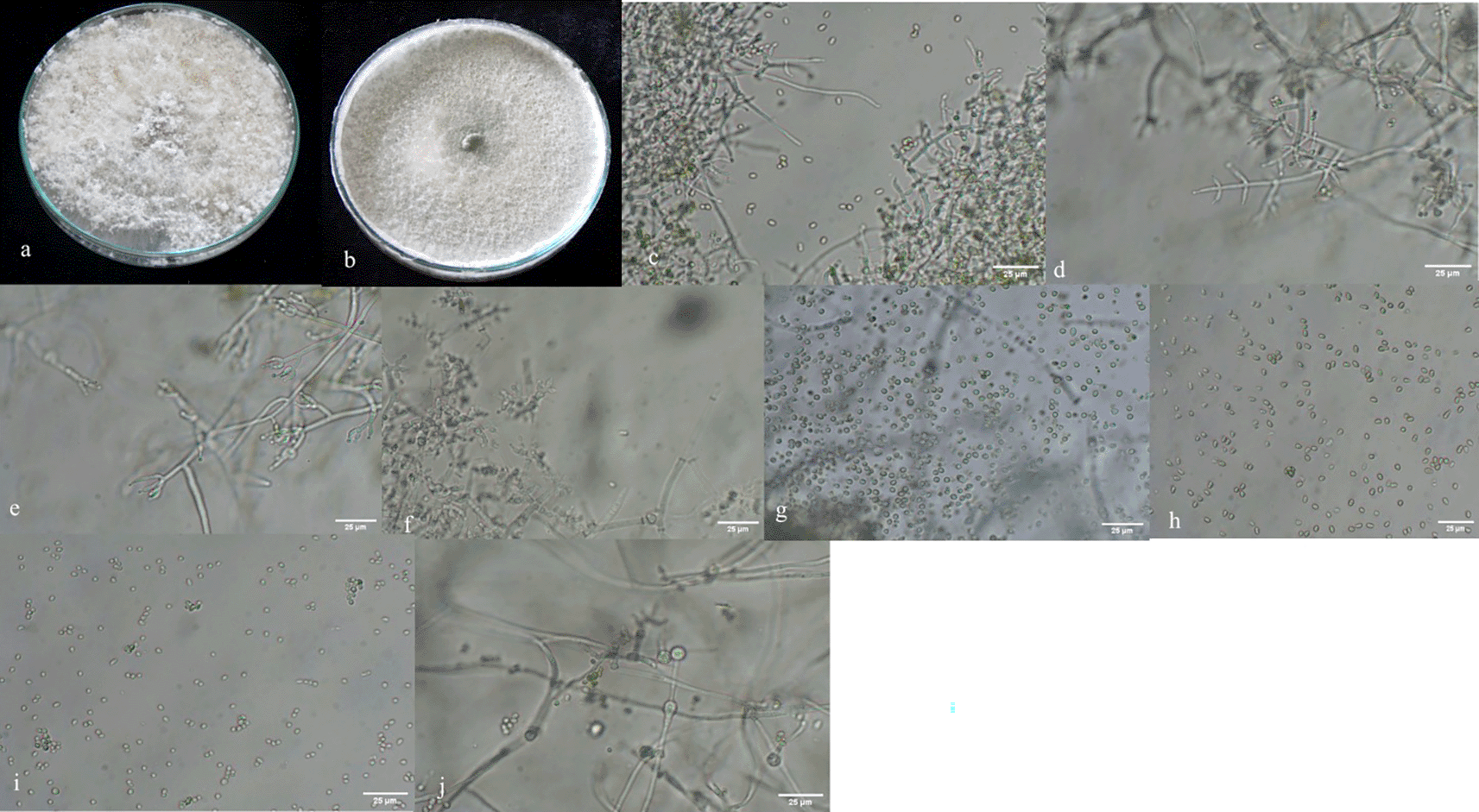
a, b: with and without aerial mycelium. c–f: different types of phialides. g–i: different types of spores. j: presence of chlamydospores in Trichoderma asperellum.
Morphological variations were observed among different groups of Trichoderma isolates (Table 1). Illustrations of colonies grown in PDA Petri plates are shown in Plate 1.70 Various diffusible pigments were observed, displaying different colors, which were compared to the Audrey & Bear Color Chart (https://www.audreyandbear.com). In the Harzianum clade, which includes species such as T. afroharzianum, T. lentiforme, T. camerunense, T. inhamatum and T. azevedoi colonies exhibited 1-2 concentric rings with the production of green-colored conidia. No diffusible pigments were detected in PDA medium, and a coconut odor was also not noticeable. The conidia were oval to oblong and ranged from 0.6-4.1μm. Within the Viride clade, comprising species like T. asperellum, T. asperelloides, T. koningii and T. atroviride colonies on PDA displayed a somewhat granular appearance with green and white regions distributed throughout the plate. No diffusible pigments were detected in the PDA medium, but coconut odor was found in some groups and absent in others (Table 1). The conidia were generally round and ranged from 1-4 μm. The T5 and T17 groups of isolates displayed white mycelium with green pustules and produced yellow diffusible pigments, classifying them within the Longibrachiatum clade. Conidia in this group were oblong and ranged from 2-3.9 μm. Isolates from T20 group were challenging to differentiate and were grouped together, with conidia measuring 1.2-2.2μm. Phialides were flask-shaped in both the Harzianum and Viride clades and were present in whorls in almost all species, except in ST3 (T. afroharzianum) and ToT11(T. longibrachiatum), where they were solitary.
The Harzianum clade was found within an altitudinal range of 80-1800 masl, spanning from lowland plains to hilly regions, and was common in both tropical and temperate regions (Table 2). Similarly, the Viride clade were also identified in both tropical and temperate regions, with altitudes ranging from between 90-1475 masl, which was generally lower than that of the Harzianum clade. Isolates of the T20 group (T. brevicompactum) were primarily located in lowland plains (at 80 masl), rather than in higher mountain region, whereas T. longibrachiatum was primarily found in mid-hill regions, with altitudes ranging from 1300-1500 masl (Table 2). However, morphological characteristics alone proved insufficient for distinguishing between the various Trichoderma isolates. Therefore, molecular identification was required to differentiate among complex and overlapping Trichoderma isolates.
Trichoderma isolates were further identified using molecular sequencing data of ITS rRNA and tef-1α genes. Out of 33 strains, PCR and bidirectional sequencing were successfully completed for 29 presumptive isolates by ITS oligonucleotide barcode identification (OR140790-OR140818) and for only of 26 isolates using tef-1α (OR567133-OR567158). The average consensus sequence length for tef-1α was 663 bp, while for the ITS rRNA, it was 646 bp. Nucleotide BLAST analysis and pairwise alignments of the ITS rRNA and tef-1α sequences identified the Trichoderma isolates as belonging to T. harzianum, T. afroharzianum, T. lentiforme, T. inhamatum, T. camerunense, T. azevedoi, T. atroviride, T. asperellum, T. asperelloides, T. brevicompactum, T. koningii and T. longibrachiatum which shared 97-100% identity match with published sequences in the NCBI database.
The phylogenetic tree (Figure 4) based on ITS rRNA unambiguously identified 11 species among the 29 isolates, based on their clustering with reference taxa. The isolates identified in the analysis included T. harzianum, T. afroharzianum, T. lentiforme, T. inhamatum, T. camerunense, T. azevedoi, T. asperellum, T. asperelloides, T. brevicompactum, T. koningii and T. longibrachiatum. Although the ITS rRNA tree could not clearly delimit the species resolution of the isolates MT8, ST49, MT5, and PanT1, however there were clear and distinct resolution of other isolates (Figure 4). The phylogenetic tree (Figure 5) obtained from the tef-1α region identified 10 species among the 26 isolates, which included T. afroharzianum, T. lentiforme, T. inhamatum, T. camerunense, T. azevedoi, T. atroviride, T. asperellum, T. asperelloides, T. longibrachiatum and T. brevicompactum. The maximum likelihood phylogenetic tree, based on the concatenated dataset ITS and tef-1α (1352 bp), produced four distinct, well supported clades with bootstrap values higher than 70%: Harzianum, Viride, Longibrachiatum and Brevicompactum clades (Figure 6).
Thus, based on above three phylogenetic trees, four clades were identified: Harzianum, Viride, Brevicompactum and Longibrachiatum clades. Eighty-five isolates belonged to the Harzianum clade, consisting of T. afroharzianum (64 isolates), T. harzianum (3 isolates), T. lentiforme (1 isolate), T. camerunense (11 isolates T. inhamatum (5 isolates), and T. azevedoi (1 isolate). The Viride clade included 67 isolates, comprising species such as T. asperellum (57 isolates), T. asperelloides (6 isolates), T. atroviride (1 isolate), and T. koningii (3 isolates). The Longibrachiatum clade comprised 11 isolates, all belonging to the T. longibrachiatum. The Brevicompactum clade consisted of 4 isolates, all of which were identified as T. brevicompactum (Table 3). Among these, T. afroharzianum was the most abundant species in this study, followed by T. asperellum.
Analysis of the diversity among Trichoderma species
Table 4 shows Simpson’s biodiversity index (D), Shannon’s biodiversity index (H), evenness (J), and the abundance index (E) for various agricultural fields in different ecological zones. The highest species diversity and evennesswere recorded in the high mountain region, with Shannon’s index (H) of 1.724, evenness (J) of 0.84 and Simpson’s index (D) of 0.28). Following this the hilly region which exhibited the Shannon’s index of 1.563, eveness of 0.7 and Simpson’s index of of 0.28. In contrast, the plain region had the lowest species diversity with Shannon and Simpson diversity indices and evenness estimatedto be H = 1.515, J=0.66, D = 0.33. The species abundance values were E = 2.11 for the plain, E = 1.95 for mid-hill and E = 1.99 for the high mountain region. The dominance values (Y) of T. arfoharzianum T. asperellum, T. camerunense and T. longibrachiatum were 0.04, 0.03, 0.01, and 0.01 respectively and they were classified as the principal species. On average, the diversity values for Trichoderma species were H = 1.61, D = 0.29, J = 0.74, and E= 2.02 (Table 4). Simpson’s index (1-D) and the evenness index were close to 1 in all regions except for the plain region, indicating a very high diversity of Trichoderma species in the major agricultural areas of Nepal. The number of species and isolates, as well as the dominant species of Trichoderma, varied geographically (Table 5) revealing that the mid hill and high mountain regions have a high diversity of Trichoderma species.
The remarkable biodiversity found in Nepal can be attributed directly to its unique geographical position, which spans a broad spectrum of altitudinal variances and a wide array of climatic conditions. Consequently, Nepal holds a significant position as one of the world’s important biodiversity hotspots. The fungal genus Trichoderma has been extensively studied for its diverse metabolites and their applications in agriculture, industry and health.6 Its ability to establish beneficial interactions with plants and produce bioactive compounds makes it a valuable resource for sustainable and eco-friendly solutions in multiple sectors.26,27,54 However, there have been very few studies on the diversity of Trichoderma species in Nepal compared to other parts of the world.10,15,35
In the present study, the isolates of Trichoderma (n = 167) isolated from soils of different ecological zones were categorized into three clades based on their morphological characterization: Harzianum, Viride and Longibrachiatum. However, some isolates were challenging to differentiate and were not assigned to any specific clade (T20). Our observations in terms of morphological characterization align with the previous studies on these species.38,55 The use of a single method for identifying Trichoderma species can present challenges and potential issues. To address this concern, Bissett proposed integration of both morphological and molecular studies.56 This combined approach allows for a more comprehensive and accurate species-level classification within Trichoderma spp. In our study, species were identified following previous studies that utilized ITS and tef-1α sequencing.57 Using molecular approaches, we were able to clearly identify some cryptic species, and we also distinguished isolates that could not be identified solely based on morphological features.
Trichoderma species are typically identified through DNA gene sequence analysis, with the generally accepted practice being the use of two gene sequences. However, recent studies suggest that the identification of new Trichoderma species may require the analysis of at least three DNA gene sequences.58,59 In this particular study, 33 Trichoderma isolates were selected from a pool of 167 isolates isolated from soil samples that were collected from various regions of Nepal (Figures 1 and 2). To identify these isolates, we analyzed two DNA gene sequences: the ITS rRNA and the tef-1α genes. This analysis involved comparing the obtained sequences with those available in the NCBI database using BLASTn analysis. It has been observed that relying solely on individual gene sequences, such as ITS rRNA or tef-1α , is insufficient for accurately distinguishing all Trichoderma species. As a result, we used a combination of ITS rRNA and tef-1α gene sequences, for a more comprehensive understanding of Trichoderma species distribution. In BLASTn searches, ITS sequences did not provide sufficient resolution of Trichoderma phylogeny for some species, such as the T. harzianum complex. However, the concatenated dataset (ITS-tef-1α ) produced four well-supported clades. The resulting tree grouped together the same or closely related species into distinct clades (Figure 2). The comparison of test and conference taxa aligned with earlier reports.12,60,61 The findings revealed similarities among Trichoderma species, with the out-group sequence of Fusarium oxysporum forming dissimilar clusters. Although tef-1α sequencing provided a clear picture of Trichoderma phylogeny, the predominant species complex (T. harzianum complex) comprises multiple cryptic species.62,63 The T. harzianum complex has undergone significant revision in recent times, resulting in the classification of T. harzianum into 14 distinct species.64,65
In this research, we conducted a comprehensive examination of the diversity of indigenous Trichoderma species found in the rhizospheric soil across different districts of Nepal, taking into account variations in altitude. We assessed Trichoderma species diversity, which is defined as the product of the evenness and the number of species, using the Shannon biodiversity index -H.51 To evaluate the dominance of individual species, we calculated Simpson’s diversity index,50 which indicates the probability that two randomly selected species from a given ecosystem will belong to different categories. We also used Margalef’s abundance index to assess species richness and the Pielou index to determine the evenness of the Trichoderma population. The diversity and occurrence of Trichoderma species reported from different agroclimatic zones of Nepal, namely, the plain (80-141 masl), mid-hill (1284-1650 masl), and high mountain (1800-2500 masl) regions, clearly indicate that climatic topography and soil type are major factors influencing the distribution of Trichoderma species.
The present study aimed to assess the suitably of several widely used diversity indices for various types of statistical analyses. We conducted both simple and multifaceted statistical analyses to determine if certain indices were better suited for specific types of analyses. Notably, Simpson’s index(1-D) and the evenness index approached 1 in the mid-hill and high mountain regions, except in the plain region. This observation suggests a remarkably high diversity of Trichoderma species in the major agricultural areas of Nepal, particularly in the mid-hill and high mountain regions. This is obvious that mountains and hills have high climatic diversity due to latitudinal variation compared to plain and harbor higher diversity of Trichoderma species compared to plain.
These findings underscore the substantial diversity of Trichoderma species in these specific regions. Interestingly, Shannon’s index yielded quite similar values (~1.5) in the plain and mid-hill regions, which could be attributed to the extensive disturbance caused by human activities in both regions. The results of this study indicate that Trichoderma spp. diversity and habitat preference can serve as natural indicators of rhizosphere soil health. Moreover, our findings align with the research conducted by Ma49 in Chinese grasslands of China and Mulatu66 in Ethiopian coffee ecosystems. This diversity analysis of Trichoderma strains will facilitate the improved identification of Trichoderma species with biocontrol mechanisms, which can, in turn, contribute to the development of suitable bioformulations in sustainable agriculture.
The number of Trichoderma species and isolates, as well as the dominant species, varied significantly across different geographical regions. The plain region (80-141 masl) encompasses two districts, Sunsari and Morang. In this region, we identified a total of six different species of Trichoderma: T. afroharzianum, T. lentiforme, T. inhamatum, T. asperellum, T. brevicompactum and T. azevedoi. We found a total of 72 isolates in the plain region, with T. afroharzianum (38 isolates) being the dominant species. The mid-hill region (1284-1650 masl) includes four main districts: Kathmandu, Lalitpur, Bhaktapur, and Kavrepalnchok. Here we identified six different types of Trichoderma species: T. afroharzianum, T. camerunense, T. longibrachiatum, T. asperellum, T. asperelloides and T. atroviride with total of 61 isolates. In the mid-hill region, T. asperellum (26 isolates) was the dominant species. The high mountain region (1800-2500 masl) comprises two districts, Solukhumbu and Sindhupalchok. In this region, we found only four Trichoderma species: T. harzianum, T. afroharzianum, T. asperellum and T. koningii, with a total of 34 isolates. In this region, T. asperellum (16 isolates) was the dominant species. The presence of these Trichoderma species in the rhizospheric soils of vegetables in Nepal can be attributed to the diverse ecological substrates and climate conditions in the country’s vegetable crop growing areas. Our results suggest that these areas provide an optimal environment for the survival and colonization of a diverse group of Trichoderma species. Trichoderma afroharzianum (39.0%) was found to be the most widely distributed and abundant species in our study. The occurrence of Trichoderma species is influenced by various factors, including metabolic diversity, reproductive capabilities, substrate availability and the competitive abilities of Trichoderma isolates in natural ecosystems.
In the present, we identified nine new country record of Trichoderma species: T. afroharzianum T. harzianum, T. lentiforme, T. inhamatum, T. camerunense, T. atroviride, T. brevicompactum, T. longibrachiatum and T. azevedoi. However, these species have previously been described in regions across the world, including America,63,67,68Asia,10,15,16Africa9,31,66and European Mediterranean countries.23,42 During this study, we reisolated some species, including T. asperellum, T. asperelloides and T. koningii, along with nine additional species. However, two previously reported species, T. rugulosum and T. lixii were not found in this study.10,35 The Harzianum clade contained economically important species such as T. harzianum, T. afroharzianum, T. camerenunse, T. lentiforme and T. inhamatum, which are used in agriculture as biological control agents. The Viride clade included T. asperellum, T. asperelloides, T. koningii and T. atroviride. The Longibrachiatum clade consisted of T. longibrachiatum, while the Brevicompactum clade with T. brevicompactum. T. longibrachiatum has high optimal and maximum growth temperatures and exhibits yellow reverse pigmentation due to the production of secondary metabolites such as pyrone. T. brevicompactum is utilized in the production of various antimicrobial substances, offering significant agricultural, health, and environmental benefits.
In this study, we obtained isolates of T. harzianum, T. afroharzianum, T. inhamatum, T. lentiforme, T. azevedoi, T. camerunense, T. asperellum, T. asperelloides, T. atroviride, T. koningii, T. longibrachiatum, and T. brevicompactum from various crop ecosystems. Notably, T. afroharzianum and T. asperellum were most widely distributed species in our study. However, previous studies in crop ecosystems in Nepal have reported T. asperellum and T. asperelloides as the most widely distributed species of this genus.35 It is worth noting that among these reported species, only T. asperellum, T. asperelloides and T. koningii have been previously documented in Nepal.10,35
This study presents novel findings regarding the presence of Trichoderma species in the Himalayan foothills of Nepal at different altitudes in previously unexplored geographic regions. However, it is important to note that our observations provide only a limited glimpse into the overall diversity of Trichoderma in Nepal. Despite being recognized as a biodiversity hotspot, fungal diversity in this region remains poorly understood. To gain a more comprehensive understanding, further investigations involving the isolation of Trichoderma from various substrates like forest areas and high-altitude areas in Nepal are highly recommended. Such endeavors are likely to unveil a multitude of additional species, some of which may be previously unknown. These new discovered taxa hold significant potential for various biotechnological applications and can contribute to further advancements in this field.
Authors and Affiliations
Puja Jaiswal
Central Department of Zoology, Tribhuvan University, Kirtipur, Nepal
Ram B. Khadka and Suraj Baidya
National Plant Pathology Research Center, Nepal Agricultural Research Council, Khumaltar, Nepal
Aashaq Hussain Bhat
Department of Bioscience, University Centre for Research and Development, Chandigarh
University, Punjab, India
Arvind Kumar Keshari
Department of Zoology, Patan Multiple Campus, Tribhuvan University, Lalitpur, Nepal
Puja Jaiswal conceptualized, did data curation, formal analysis, investigation, methodology, validation and writing original draft. Ram Bahadur Khadka worked on conceptualization, formal analysis, investigation, methodology, project administration, resources, supervision, and validation of the study. Suraj Baidya helped with writing, review and editing the manuscript. Aashaq Hussain Bhat did formal analysis, validation and writing, review and editing the manuscript. Arvind Kumar Keshari conceptualization, project administration, resources, supervision, validation, writing, review and editing the manuscript. All authors read and agree to the final draft of the paper.
The present study didn’t involve the use of human and animals so ethical approval and consent were not required.
The sequencing data are available on the NCBI Genbank webpage
Morphological and Molecular Characterization of Native Trichoderma isolates from Nepal (OR140790- OR140818; OR567133-OR567158)
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140790; https://www.ncbi.nlm.nih.gov/nuccore/OR140790 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140791; https://www.ncbi.nlm.nih.gov/nuccore/OR140791 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140792; https://www.ncbi.nlm.nih.gov/nuccore/OR140792 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140793; https://www.ncbi.nlm.nih.gov/nuccore/OR140793 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140794; https://www.ncbi.nlm.nih.gov/nuccore/OR140794 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140795; https://www.ncbi.nlm.nih.gov/nuccore/OR140795 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140796; https://www.ncbi.nlm.nih.gov/nuccore/OR140796 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140797; https://www.ncbi.nlm.nih.gov/nuccore/OR140797 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140798; https://www.ncbi.nlm.nih.gov/nuccore/OR140798 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140799; https://www.ncbi.nlm.nih.gov/nuccore/OR140799 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140800; https://www.ncbi.nlm.nih.gov/nuccore/OR140800 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR1407801; https://www.ncbi.nlm.nih.gov/nuccore/OR1407801 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR1407802; https://www.ncbi.nlm.nih.gov/nuccore/OR1407802 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR1407803; https://www.ncbi.nlm.nih.gov/nuccore/OR1407803 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR1407804; https://www.ncbi.nlm.nih.gov/nuccore/OR1407804 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR1407805; https://www.ncbi.nlm.nih.gov/nuccore/OR1407805 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR1407806;https://www.ncbi.nlm.nih.gov/nuccore/OR1407806 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR1407807; https://www.ncbi.nlm.nih.gov/nuccore/OR1407807 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR1407808; https://www.ncbi.nlm.nih.gov/nuccore/OR1407808 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140809; https://www.ncbi.nlm.nih.gov/nuccore/OR140809 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140810; https://www.ncbi.nlm.nih.gov/nuccore/OR140810 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140811; https://www.ncbi.nlm.nih.gov/nuccore/OR140811 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140812; https://www.ncbi.nlm.nih.gov/nuccore/OR140812 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140813; https://www.ncbi.nlm.nih.gov/nuccore/OR140813 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140814; https://www.ncbi.nlm.nih.gov/nuccore/OR140814 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140815; https://www.ncbi.nlm.nih.gov/nuccore/OR140815 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140816; https://www.ncbi.nlm.nih.gov/nuccore/OR140816(Jaiswal et al. 2024)
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140817; https://www.ncbi.nlm.nih.gov/nuccore/OR140817 69
NCBI: Trichoderma Internal Transcribed Spacer RNA (ITS): Accession number OR140818; https://www.ncbi.nlm.nih.gov/nuccore/OR140818 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1): Accession number OR567133; https://www.ncbi.nlm.nih.gov/nuccore/OR567133 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567134; https://www.ncbi.nlm.nih.gov/nuccore/OR567134 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567135; https://www.ncbi.nlm.nih.gov/nuccore/OR567135 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567136; https://www.ncbi.nlm.nih.gov/nuccore/OR567136 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567137; https://www.ncbi.nlm.nih.gov/nuccore/OR567137 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567138; https://www.ncbi.nlm.nih.gov/nuccore/OR567138 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567139; https://www.ncbi.nlm.nih.gov/nuccore/OR567139 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567140; https://www.ncbi.nlm.nih.gov/nuccore/OR567140 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567141; https://www.ncbi.nlm.nih.gov/nuccore/OR567141(Jaiswal et al. 2024)
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567142; https://www.ncbi.nlm.nih.gov/nuccore/OR567142 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567143; https://www.ncbi.nlm.nih.gov/nuccore/OR567143 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567144; https://www.ncbi.nlm.nih.gov/nuccore/OR567144 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567145; https://www.ncbi.nlm.nih.gov/nuccore/OR567145 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567146; https://www.ncbi.nlm.nih.gov/nuccore/OR567146 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567147; https://www.ncbi.nlm.nih.gov/nuccore/OR567147 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567148; https://www.ncbi.nlm.nih.gov/nuccore/OR567148 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567149; https://www.ncbi.nlm.nih.gov/nuccore/OR567149 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567150; https://www.ncbi.nlm.nih.gov/nuccore/OR567150 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567151; https://www.ncbi.nlm.nih.gov/nuccore/OR567151 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567152;https://www.ncbi.nlm.nih.gov/nuccore/OR567152 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567153; https://www.ncbi.nlm.nih.gov/nuccore/OR567153 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567154; https://www.ncbi.nlm.nih.gov/nuccore/OR567154 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567155; https://www.ncbi.nlm.nih.gov/nuccore/OR567155 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567156;https://www.ncbi.nlm.nih.gov/nuccore/OR567156 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1v): Accession number OR567157; https://www.ncbi.nlm.nih.gov/nuccore/OR567157 69
NCBI: Trichoderma DNA sequence of translation elongation factor 1α (tef-1α): Accession number OR567158; https://www.ncbi.nlm.nih.gov/nuccore/OR567158 69
Figshare: Morphological and molecular characterization of Trichoderma isolates from vegetable crop rhizospheres in Nepal; https://doi.org/10.6084/m9.figshare.26380360.v1 69
Data are avalilable under the terms of the Creative Commons Attribution 4.0 Internasional license (CC-BY 4.0)
Figshare: Morphological and molecular characterization of Trichoderma isolates from vegetable crop rhizospheres in Nepal; Plate 1. Culture of Trichoderma isolates grown in 28±2°c in potato dextrose agar medium; https://doi.org/10.6084/m9.figshare.26405059.v1 70
Data are avalilable under the terms of the Creative Commons Attribution 4.0 Internasional license (CC-BY 4.0)
Authors would like to acknowledge NARC-National Plant Pathology Research Center for providing laboratory facilities, and members of NARC- Molecular Plant Pathology Lab for the support to conduct the research.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Mycology, Biocontrol
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Korkom Y, Yıldız A: First report of Trichoderma guizhouense isolated from soil in Türkiye. Journal of Plant Diseases and Protection. 2024; 131 (2): 619-625 Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Trichoderma, plant pathology, biological control, soil-borne diseases, fruit diseases, endophytic fungi, fungal pathogens,
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 3 (revision) 05 Mar 25 |
|||
|
Version 2 (revision) 09 Jan 25 |
read | read | |
|
Version 1 24 Sep 24 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)