Keywords
Conservation, complete chloroplast genome, Lauraceae, phylogenetic
This article is included in the Nanopore Analysis gateway.
This article is included in the Genomics and Genetics gateway.
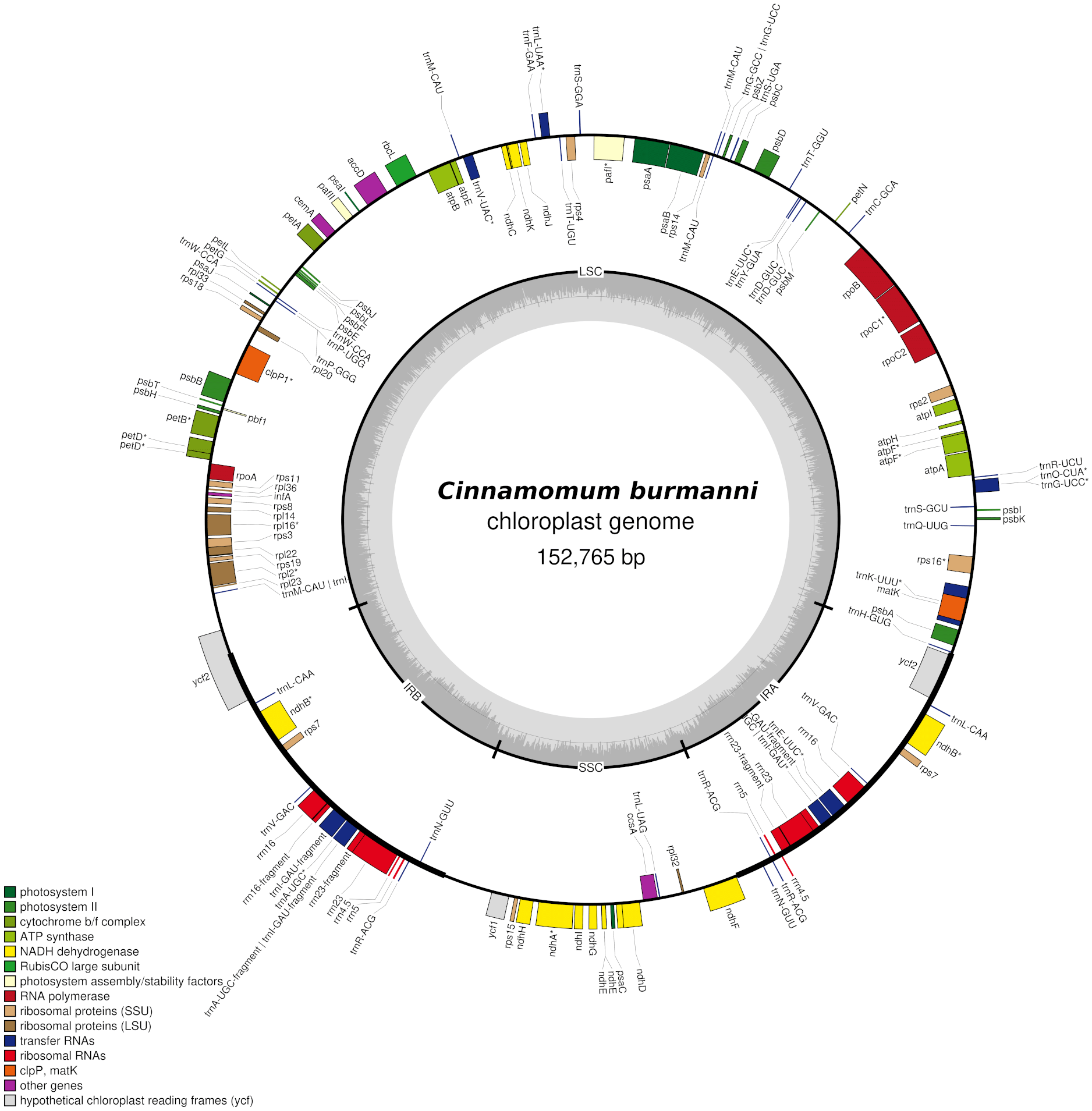
Cinnamomum burmanni (Nees & T.Nees) Blume (Lauraceae) belongs to the Magnoliids group and is mainly distributed in Indonesia and Southeast Asia. The complete chloroplast (cp) genome of C. burmanni sampled from Indonesia was assembled and annotated for the first time in this study. The length of the cp genome is 152,765 bp with a GC content of 39%, and it consists of four subregions: a large single-copy (LSC) region of 93,636 bp, a small single-copy (SSC) region of 18,893 bp and two inverted repeats (IR) regions (IRA 20,121 bp; IRB 20,115 bp) . The cp genome of C. burmanni encodes a total of 173 unique genes, which are 96 protein-coding genes, 19 rRNA genes, and 68 tRNA genes that can be utilized for advanced genetic and genomic studies of the species.
Conservation, complete chloroplast genome, Lauraceae, phylogenetic
We updated the Introduction and Result sections according to reviewers suggestion.
See the authors' detailed response to the review by Jakub Sawicki
See the authors' detailed response to the review by Hoang Dang Khoa Do
Cinnamomum burmanni (or cinnamon) is an endemic woody shrub belonging to the Lauraceae family, which is widely distributed in Indonesia, covering West Sumatra, North Sumatra, Jambi, Bengkulu, Java Island, and Maluku Islands (Suwarto et al., 2014). The species grows in altitudes between 0 and 2000 m above sea level. The tree can grow up to 15 m tall, and the wood is grey with a very distinctive aroma and sweet taste, so its wood is widely used for spices, cosmetics, and herbs. The active compounds contained in cinnamon wood are cinnamaldehyde, flavonoids, alkaloids, tannins, saponins, coumarins, steroids, eugenol, and phenols, which act as anti-bacterial, anti-tumor, antioxidant, anti-inflammatory, anti-cancer, and anti-diabetic agents (Indarto et al., 2022).
The chloroplast (cp) genome sequence of C. burmanni from China was previously elucidated by Yang et al. (2019) utilizing 11 universal primer pairs for long-range PCR in next-generation sequencing. However, their study provided only a phylogenetic tree, and the corresponding sequence data and cp genome map are not readily accessible (https://lcgdb.wordpress.com). In the present study, we present genomic resources for C. burmanni from Indonesia, generated through the long-read sequencing method utilizing MinION technology from Oxford Nanopore Technologies. This study includes a comprehensive list of genes within the chloroplast genome of C. burmanni, which was not included in the previous study. Such information is of considerable significance as a foundation for further genetic investigations. Additionaly, the study of the cp genome C. burmanni from Indonesia using PCR-free library preparation method and long-read sequencing generated by MinION Oxford Nanopore Technologies (ONT) has not been carried out, whereas this information is needed to improve correct species identification and to optimize the sustainable use of genetic resources for this species. MinION is a third-generation sequencing technology with nanopore technology (Oxford Nanopore Technologies (ONT), 2017). Furthermore, the present study aimed to re-sequence and assemble the complete cp genome C. burmanni from Indonesia using MinION Oxford Nanopore Technologies (ONT).
Fresh leaf samples were collected from one individual C. burmanni tree in Lembang Subdistrict, West Bandung Regency, West Java Province, Indonesia. The collected leaf sample was subsequently used for DNA extraction and sequencing in the field. The data analysis of the chloroplast genome was performed at the Laboratory of Forest Genetics and Molecular Forestry, Department of Silviculture, Faculty of Forestry and Environment, IPB University.
C. burmanni genomic DNA was extracted using the Qiagen DNeasy Plant Mini Kit (cat. nos. 69104 and 69106) following the manufacturer’s protocol with slight modifications. Extraction was first done by grinding fresh leaf samples to which 400 μl of Buffer AP1 and 1 μl of mercaptoethanol had been added using a mortar and pestle. The disrupted sample was placed into a 1.5 ml microcentrifuge tube. The mixture was then incubated for 10 min at 65°C using Mini Heating Dry Bath Incubator MD-MINI (Major Science Co., Ltd). Afterward, 130 μl Buffer P3 was added into the microtube and then vortexed using Biosan V-32 Multi-Vortex and incubated for 5 min on ice. The mixture was centrifuged for 2 min at 8000 × g using a portable microcentrifuge on the Bento Lab device (Bento Bioworks Ltd). The lysate was pipetted into a QIAshredder spin column placed in a 2 ml collection tube. The lysate was centrifuged for 2 min at 8000 × g using a portable microcentrifuge on the Bento Lab device (Bento Bioworks Ltd). The flow-through was transferred into a new tube without disturbing the DNA pellet. 1.5 volumes of Buffer AW1 were then added and mixed by pipetting. 650 μl of the mixture was transferred into a DNeasy Mini spin column placed in a 2 ml collection tube and subsequently centrifuged for 1 min at 6000 × g. The flow-through was discarded, and the spin column was placed into a new 2 ml collection tube. 500 μl Buffer AW2 was added to the spin column and centrifuged for 1 min at 6000 × g. The flow-through was then discarded and the spin column was transferred to a new 1.5 ml microcentrifuge tube. 50 μl Buffer AE was added for elution and subsequently incubated for 5 min at room temperature (15–25°C) before centrifuging for 1 min at 6000 × g. The quantity of genomic DNA was measured by the Invitrogen Qubit 1.0 fluorometer with the Qubit dsDNA BR assay kit.
The high molecular weight of 400 ng evaluated DNA proceeded to PCR-free library preparation, which followed the Nanopore protocol for Rapid sequencing gDNA – Field Sequencing Kit (SQK-LRK 001) version FSK_9049_v1_revR_14Aug2019. The prepared DNA library was sequenced using the MinION R9.4.1 flow cell (FLO-MIN106D) on a MinION Mk1B sequencer (Oxford Nanopore Technologies). The runs were visualized by MinKnow v3.6.5 software (http://community.nanoporetech.com Oxford Nanopore Technologies).
Basecalling with super accuracy mode (SUP) to translate Fast5 raw data into Fastq (DNA-Seq of Cinnamomum burmanni. NCBI Accession number SRX22198906) data was performed using the Guppy program (RRID:SCR_023196) v4.2.3+8aca2af8 (Wick et al., 2019). Quality control was done using NanoStat v1.5.0 and NanoPlot (RRID:SCR_024128) v1.28.2 (De Coster et al., 2018). Chloroplast genome assembly was performed using Galaxy Server (RRID:SCR_006281) version 23.1.1.dev0 (Sloggett et al., 2013). The assembly includes a filtering step to filter reads quality (Q>7) and length minimum of reads (>500 bp), using the Flye program (RRID:SCR_017016) v2.9 (Kolmogorov et al., 2020) and polishing the assembly results using the MEDAKA consensus program (RRID:SCR_005857) v1.4.4 (Oosterbroek et al., 2021). The cpDNA data was then annotated using the GeSeq (RRID:SCR_017336) on the CHLOROBOX platform (Tillich et al., 2017) to assign functions to the predicted genes and generate a map representation of the chloroplast genome.
The sequencing resulted in a total of 59,589 reads, total bases of 140,909,220 bp, and read length N50 4,377 bp. The assembly resulted in 288 contigs, contig_246 annotated as cp genome with coverage of 37x. The cp genome showed a typical quadripartite structure ( Figure 1) with a length of 152,765 bp, consisting of small single copy (SSC 18,893 bp) and large single copy (LSC 93,636 bp) regions separated by a pair of inverted repeat A (IRA 20,121 bp) and inverted repeat B (IRB 20,115 bp) regions ( Figure 1). The C. burmanni chloroplast genome contained 173 unique genes, including 96 coding sequences, 68 transfer RNA (tRNA), and 19 ribosomal RNA (rRNA) genes ( Table 1). The C. burmanni sequence had a GC content of 39% (LSC 38%; SSC 34%; IR 44%) and a coverage depth 37x. The results of the typical quadripartite structure were similar to the C. burmanni reported by Yang et al. (2019). The chloroplast genome assembly of C. burmanni has 10 bp less (152,765 bp) than previously reported by Yang et al. (2019) (152,775 bp). However, the general GC ratio of each region is comparable at 38% (LSC), 34% (SSC), and 44% (IR).
| Functional category | Group of genes | Name of genes |
|---|---|---|
| Self-replication | Large subunit ribosomal proteins | rpl32, rpl33, rpl20, rpl36, rpl14, rpl16*(×2), rpl22, rpl2*(×2), rpl23 |
| DNA dependent RNA polymerase | rpoC2, rpoC1*(×2), rpoB, rpoA | |
| Small subunit ribosomal proteins | rps7, rps12**(×2), rps15, rps7, rps16*(×2), rps2, rps14, rps4, rps18, rps11, rps8, rps3, rps19, rps12-fragment* | |
| rRNAs | rrn16, rrn23, rrn5, rrn4.5 | |
| tRNAs | trnL-CAA, trnV-GAC, trnE-UUC, trnA-UGC *(×2), trnN-GUU, trnL-UAG, trnN-GUU, trnR-ACG, trnR-ACG, trnI-GAU *(×2), trnV-GAC, trnL-CAA, trnH-GUG, trnK-UUU *(×2), trnQ-UUG, trnG-UCC *(×2), trnR-UCU, trnC-GCA, trnD-GUC, trnY-GUA, trnT-GGU, trnS-UGA, trnG-GCC, trnM-CAU, trnfM-CAU, trnS-GGA, trnT-UGU, trnL-UAA *(×2), trnF-GAA, trnV-UAC *(×2), trnW-CCA, trnP-UGG, trnI-CAU, trnS-GCU | |
| Subunit of ATP synthase | atpA, atpF*(×2), atpH, atpI, atpE, atpB | |
| Subunit of NADH-dehydrogenase | ndhB*(×2), ndhH, ndhA*(×2), ndhI, ndhG, ndhE, ndhD, ndhF, ndhJ, ndhK, ndhC | |
| Photosynthesis | Subunits of cytochrome b/f complex | petN, petA, petL, petG, petB*(×2), petD*(×2) |
| Subunits of photosystem I | psaC, psaB, psaA, psaI, psaJ | |
| Subunits of photosystem II | psbK, psbI, psbM, psbD, psbC, psbZ, psbJ, psbL, psbF, psbE, psbB, psbT, psbH, psbA | |
| Subunit rubisco | rbcL | |
| Photosystem assembly factors | pafI**(×2), pafII | |
| Photosystem biogenesis factor | Pbf1 | |
| Subunit of acetyl-CoA-carboxylase | accD | |
| C-type cytochrome synthesis gene | ccsA | |
| Other functions | Envelope membrane protein | cemA |
| ATP-dependent protease subunit P | clpP1**(×2) | |
| Maturase | matK | |
| Conserved open reading frames | ycf1 (×2), ycf2 (×2) | |
| Transitional initiation factor | infA |
NCBI’s Short Read Archive (SRA): DNA-Seq of Cinnamomum burmanni. Accession number SRX22198906; https://www.ncbi.nlm.nih.gov/sra/SRX22198906 (IPB University, 2023).
The authors thank Mrs. Siska Nurfajri and Ms. Nabila Salsabila Salmah, the alumni of the Department of Silviculture, Faculty of Forestry and Environment, IPB University, for their valuable assistance in DNA extraction.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bioinformatics, Genetics, Biotechnology
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Entomologist, Evolutionary Biology
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genomics, Chloroplast genome evolution, Phylogeny
Are the rationale for sequencing the genome and the species significance clearly described?
No
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: genome
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Entomologist, Evolutionary Biology
Are the rationale for sequencing the genome and the species significance clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
Partly
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bioinformatics, Genetics, Biotechnology
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genomics, Chloroplast genome evolution, Phylogeny
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Chloroplast genomics, nanopore sequencing
Are the rationale for sequencing the genome and the species significance clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
Partly
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Chloroplast genomics, nanopore sequencing
Are the rationale for sequencing the genome and the species significance clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genomics, Chloroplast genome evolution, Phylogeny
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
|
Version 3 (revision) 16 Dec 25 |
read | read | read | read | |
|
Version 2 (revision) 21 May 24 |
read | read | read | read | read |
|
Version 1 20 Feb 24 |
read | read | |||
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)