Keywords
Chronic Myeloid Leukemia, High-Performance Liquid Chromatography, Polymerase chain reaction, Restriction fragment length Polymorphism
This article is included in the Oncology gateway.
To ascertain the impact of CYP3A4 polymorphism on the therapeutic outcome of imatinib and its trough concentration
Descriptive analytical study
This study was conducted in the Department of Pharmacology & Therapeutics, Islamic International Medical College, between December 2020 and February 2022, in collaboration with the Institute of Biomedical Genetic Engineering, KRL Hospital Islamabad.
Patients with Chronic Myeloid Leukaemia age range–18-70 years were included in this study. One group comprised responders and the other group comprised non-responders. Imatinib trough levels in both groups were determined using High-performance Liquid Chromatography and compared, and the association was determined with therapeutic outcomes. DNA was extracted, and the PCR restriction fragment length polymorphism technique was used to identify the alleles. The results were analyzed using Statistical Package for Social Sciences (SPSS) version 22.0.
The imatinib concentration in patients with the homozygous wild allele CC of rs2242480 was higher(1298ng/ml) as compared with the mutant homozygous TT allele (489ng/ml) or heterozygous allele CT(873ng/ml)
There was a significant association between imatinib trough levels and CYP3A4 polymorphism, and trough concentration was found to be lower in patients with the TT or CT variant of rs2242480 than in patients with the CC genotype.
Chronic Myeloid Leukemia, High-Performance Liquid Chromatography, Polymerase chain reaction, Restriction fragment length Polymorphism
The prognosis of Chronic Myeloid Leukaemia (CML) has changed significantly in the last two decades with the introduction of tyrosine kinase inhibitors (TKIs), such as imatinib mesylate (IM), as the standard treatment. The efficacy of IM is determined by its concentration in the plasma. However, there is a noticeable variability in the response among individuals, and even within the same patient, when blood samples are taken at different times. This variability in drug response could be due to changes in pharmacokinetic processes, along with other factors (Natarajan, Kumar et al. 2019). Single nucleotide polymorphisms (SNPs) or single nucleotide variants (SNVs) may affect the plasma drug concentration and hence the therapeutic response. Genes that encode transporters and metabolizing enzymes are of special interest as SNP for these enzymes may explain the resistance developed against Imatinib Mesylate in CML initially or during treatment. The cytochrome P450 enzyme system extensively metabolizes imatinib in pharmacokinetic processes (Pena, Muriel et al. 2020). The main enzyme involved in vivo metabolism is CYP3A4. Multiple studies have indicated that individuals with minor alleles of SNP rs2242480 of CYP3A4 may have lower plasma concentrations (Pena, Muriel et al. 2020). Considering this, a decreased response to treatment in individuals with minor allele frequencies could be anticipated (Ullah, Ali et al. 2022). The predicted response can lead to the adoption of strategies like therapeutic drug monitoring for Imatinib with individualized adjustment of doses to achieve the therapeutic goal during the early stages of treatment (García-Ferrer, Wojnicz et al. 2019).
This study aimed to explore the impact of minor allele frequencies of rs2242480 on the response to imatinib and plasma trough concentration with the effects of different genetic models on the outcome of treatment.
It was a descriptive analytical study and was conducted in the Department of Pharmacology and Therapeutics at Islamic International Medical College from December 2020 to February 2022.
This study is approved by Islamic International medical college Research Ethics committee, which is in alignment with the international guidelines provided by the declaration of Helsinki and International conference on harmonization –Good Clinical practice (ICH-GCP). The date of approval is July 25th, 2019, and the approval number is Ref#Riphah/IIMC/ERC/19/0265. With the necessary approval from Research Ethics Committee, data collection was started after the informed consent is taken from the participants with no financial burden on the study participants. Study participants are free to access the data and has the right to withdraw themselves from the study at any stage. As this study is observational, so direct drug intervention or therapy was not done by the researcher and no risk or harm is involved with this study. The Declaration of Helsinki was followed in maintaining the confidentiality of the study’s data.
Newly diagnosed patients were selected from the CML clinic at Holy Family Hospital. The inclusion criteria were newly diagnosed patients who were taking imatinib 400 mg daily (Haque, Shah et al. 2022), had no other comorbidities, showed good compliance with treatment, and were not receiving CYP3A4 or CYP3A5 inhibitors or inducers (Sacha 2014). Sampling technique was nonprobability consecutive sampling. Initially sample size (https://csg.sph.umich.edu/abecasis/cats/gas_power_calculator/index.html) was 120 patients, but later, due to non-compliance and findings of comorbidities, the final sample size was 106. The age of these patients ranged from to 18-70 years. Informed written consent was obtained from all participants before the start of the study.
Leukaemia net guidelines were followed to determine the treatment prognosis (Hochhaus, Baccarani et al. 2020). In some patients complete haematological response was achieved after three months of treatment with Imatinib and they were labelled as responders to the treatment of TKIs more specifically to Imatinib (SULTAN, JAFFRI et al. 2021). These patients were followed for the next three months as well to confirm their response towards the same dose of Imatinib. In patients who were labelled as non-responders, the complete haematological response could not be achieved after three months of 400 mg of Imatinib, they were Philadelphia positive as well. Therefore, they were switched to the second generation of the tyrosine kinase inhibitor nilotinib.
Genomic DNA was extracted from the blood using a Thermo Scientific Gene JET Whole Blood Genomic DNA Purification Mini Kit (Thermo Fisher Scientific Inc. Pittsburgh, PA, USA) according to the manufacturer’s protocol (Mumtaz, Ahmed et al. 2020). This method has been previously used with excellent DNA yield of good quality (Al-Saud, Al-Romaih et al. 2020). The genetic polymorphisms of CYP3A4 rs2242480 were determined by using the polymerase chain reaction (PCR) restriction fragment length polymorphism (RFLP) method (Ozeki, Nagahama et al. 2019). LEFT PRIMER: GAGGTTTTTTACTTAGCAT RIGHT PRIMER: CCCAGTGTACCTCTGAAT and the product size was 554.
When the IM plasma concentration reached a steady state, which was achieved by regularly taking IM for at least a month, peripheral blood samples were taken 0.5 hours before the IM dosing (Chen, Dong et al. 2020). These samples were analyzed for IM trough plasma concentration determination and genotyping analysis. The analysis was performed using reverse-high-performance liquid chromatography. The equipment used was a Waters e2695 separation module, along with a 2489 UV/Vis detector and autosampler. A C18 column with a particle size of 5 μm was employed, and the analysis time was 10 min at a flow rate of 1 ml/min. The mobile phase consisted of an aqueous buffer solution of KH2PO4 (0.094 M), K2HPO4 (0.0058 M), and acetonitrile at a v/v ratio of 90:10. The detector wavelength was set at 210 nm (Boovizhikannan, Mahesh et al. 2022).
Statistical analyses were conducted using SPSS software versions 22.0 and Microsoft SPSS-23 (https://www.ibm.com/support/pages/downloading-ibm-spss-statistics-22). Descriptive statistics were used to present the mean ± SEM in groups. The chi-square test was used to compare categorical variables, and the Hardy-Weinberg equilibrium test was performed using an appropriate χ2 test (Zhang, Ge et al. 2022). Binary logistic regression analysis was carried out to determine the impact of genotypes on response, and the association between genotypes was compared. Imatinib levels were compared according to genotype using analysis of variance (ANOVA). Statistical significance was set at P < 0.05.
In total, 120 patients were included in this study. Of these, 14 were excluded because they did not meet the inclusion criteria. complete Haematological response was considered in responders when the WBC count was less than 10×103/μL, basophils were less than 6% with no myelocytes, promyelocytes, or myeloblasts in the differential leukocyte count, and platelets were less than 450×103/μL with non-palpable spleen (Farooq, Khan et al. 2019). 54.7% of patients were male, while 45.3% patients were female. A significant association was found between sex and increased response in females as compared to males (64% vs. 36%). The imatinib trough concentration in males was lower than that in females (875 vs. 1151ng/ml). (Of the total sample size, 11 patients were above 60 years of age). There was an increased percentage of responders with increasing age and imatinib levels of 625 ng/ml in group 1 compared to 1240 ng/ml in group 4. None of the observed genotype frequencies were significantly different from the expected value (p > 0.05), illustrating that these frequencies follow Hardy-Weinberg equilibrium (Table 1).
| SNP ID | Gene | Genotype | n | Allele | Allele frequency | HWE p value |
|---|---|---|---|---|---|---|
| rs2242480 | CYP3A4 | CC | 49 (46.2 %) | C | 136(64 %) | 0.07 |
| CT | 38 (35.8 %) | T | 76(36 %) | |||
| TT | 19 (17.9 %) | |||||
| Total | 106 | 212 |
Of the 106 sample sizes, 49 patients had a CC variant of CY3A4 rs2242480, 38 patients had a CT variant, and 19 patients had a minor TT allele. Plasma HPLC levels of trough concentration of imatinib were calculated in all three variants with the highest trough levels of 1298 ng/ml in patients with the CC variant, while CT levels were 873ng/ml and TT levels were 489 ng/ml (Figure 1). Analysis of variance was used to determine the difference between the polymorphic variants, which was found to be significant (p-value 0.000) (Table 1). The p value between the wild homozygous and heterozygous individuals was 0.015. Similarly, the p-value was again found to be significant (0.003) between heterozygous (CT) and mutant homozygous (TT).
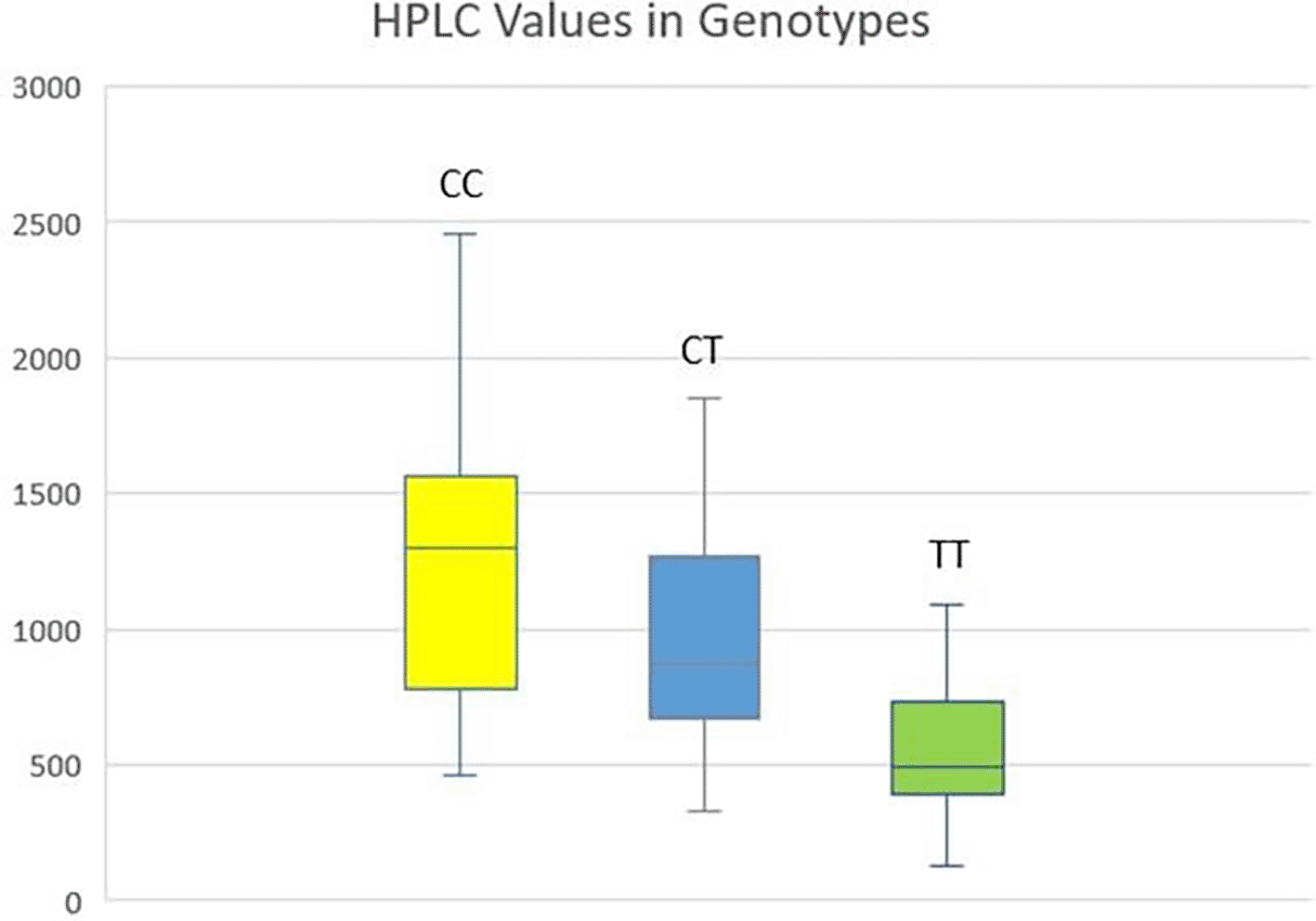
Abbreviations: HPLC: high-Performance Liquid Chromatography; CML: Chronic Myeloid Leukaemia.
CYP3A4 rs2242480 polymorphism in genetic models with impact on Imatinib concentration in non-responders and responders.
Analyzing responders and non-responders, rs2242480, minor allele TT was associated with decreased response to imatinib as a significant p-value in codominant (p-value 0.005), dominant (p value0.002), recessive (p-value 0.02), and additive models (p-value 0.000), whereas in the over-dominant model, the minor allele TT caused a decrease in response with genotypes CC/TT with a p-value of 0.199 (Table 2).
| Model | Genotype | Non Responders n = 47 | Responders n = 59 | B | OR | 95 % CI | p value | |
|---|---|---|---|---|---|---|---|---|
| Lower | Upper | |||||||
| Codominant | CC | 14 | 35 | 1 | 0.005* | |||
| CT | 20 | 18 | -1.022 | 0.360 | 0.148 | 0.875 | ||
| TT | 13 | 6 | -1.689 | 0.185 | 0.059 | 0.582 | ||
| Dominant | CC | 14 | 35 | 1 | 0.002* | |||
| CTTT | 33 | 24 | -1.235 | 0.291 | 0.129 | 0.656 | ||
| Over dominant | CCTT | 27 | 41 | 1 | 0.199 | |||
| CT | 20 | 18 | -0.523 | 0.593 | 0.266 | 1.320 | ||
| Recessive | CCCT | 34 | 53 | 1 | 0.020* | |||
| TT | 13 | 6 | -1.217 | 0.296 | 0.103 | 0.854 | ||
| Additive | C | 48 | 88 | 0.356 | 0.000* | |||
| T | 46 | 30 | -1.034 | 0.199 | 0.635 | |||
Drug transporters, including entry and efflux transporters, along with enzymes involved in imatinib metabolism, may have a substantial influence on therapeutic response. The pharmacokinetic processes of imatinib depend on the efficiency of a protein involved in drug absorption and disposition. After oral administration of IM, it is mainly metabolized by the Cytochrome P450 enzyme system by CYP3A4 and CYP3A5 enzymes. The entry transporters involved are from the ABC subfamily. Organic cationic transporter 1(OCT), encoded by SLC22A1, cannot be ignored because of its importance in the uptake and extrusion of small toxic by-products produced endogenously. Imatinib has changed the course of this cancer, but it is now widely accepted that resistance to this drug is emerging. In this study, we explored the effect of genetic polymorphisms on imatinib trough concentration determined using HPLC and its association with the outcome of treatment in responders and non-responders. This genetic polymorphism was identified against CYP3A4 rs2242480, and data were collected against each allelic form and its association with response. Sex and age associations were also assessed. Imatinib determined by HPLC was also analyzed in both sexes, in different age groups, and against each allelic form.
During the study period, from December 2020 to February 2022, blood samples were collected after steady-state concentration was achieved, and it was revealed that HPLC trough levels of imatinib were higher in the elderly than in the younger age groups, with a highly significant p-value (0.01) among groups. The trough levels of imatinib were 625 mg/ml, 780 mg/ml, 801 mg/ml, and 1240 mg/ml from group 1 to group 4, respectively, showing a gradual increase in the concentration of imatinib with increasing age. The potential mechanism behind this could be attributed to the lower volume of distribution and slower metabolism. This finding may guide treating physicians for therapeutic drug monitoring in both elderly and younger patients for dose adjustments with better treatment outcomes in terms of a decrease in adverse effects and relatively higher doses in young patients to achieve therapeutic concentration (Gao, Wang et al. 2023). Our study was in alignment with the study done in July 2023 by Adattini, Adiwidjaja et al. (2023) which showed increased concentration in the older patient population with earlier achievement of complete molecular response and more adverse effects beyond the therapeutic concentration. Another study conducted in April 2020 by Xia, Chen et al. (2020) concluded that age could be a significant covariate in determining imatinib concentration, which also supports this study for anticipating age-related adverse effects with underlying supra-therapeutic imatinib concentrations. The association of response with age was also determined through X2 and it was found to be significant (p value 0.02) with more responders to the treatment in the elderly, which aligns with the effectiveness of Imatinib in CML as determined by Chen, Dong et al. (2020). In terms of response, it was found out that females have a better response as compared to males with responder’s percentage (64% vs 36%) as shown in Table 1 and highly significant association (p value 0.000). The reason for this better outcome could be differences in pharmacokinetic processes, better immune response, or adherence to the treatment, as shown by Breccia, Molica et al. (2018). Imatinib plasma concentration was also higher in females (1151 ng/ml) vs males (875 ng/ml).
SNPs may contribute to inter-individual variations in IM reactions and disposition among different sexes. The mutant allele T was reported to be associated with a higher CYP3A4 metabolic activity, thus enhancing the clearance of IM, which is mainly metabolized by CYP3A4, leading to a substantial decrease in IM plasma levels, which was in agreement with the study done by Pena, Muriel et al. (2020). Therefore, the mutant allele T of rs2242480 is a significant risk factor for predicting inadequate clinical efficacy of IM, and patients who carry mutant allele T of rs2242480 may be suggested to have a higher dose therapy (Fava, Rege-Cambrin et al. 2021).
The results of this study were narrowed down to show not only a substantial link with the trough IM concentration, but also differential results in the same single nucleotide polymorphism. This is because the findings are all related to issues that have a strong association within their respective domains of concern. In particular, single nucleotide polymorphism of rs2242480 that showed a notable variation in IM concentration warranted or demanded more samples to confirm if they should be considered before IM administration or dose adjustment.
In conclusion, for the treatment of CML, individualized medicine requires the identification of specific SNPs linked to low imatinib concentrations. Individuals who possess these genetic variants might need to change their dosage, try different treatment plans, or be closely watched to ensure maximal therapeutic efficacy. Furthermore, by comprehending the genetic underpinnings of Imatinib metabolism, adverse drug reactions can be anticipated and managed, improving therapy overall.
Although genetic variants frequently exhibit population-specific differences, their analysis requires a diverse range of patient cohorts to account for various obstacles and restrictions. Additionally, the precise role of individual SNPs in influencing imatinib concentration is difficult to determine because of the multifactorial nature of the medication response. This response is influenced by complex interactions between genetic, environmental, and clinical factors.
Future studies should focus on expanding our knowledge of the genetic factors that influence imatinib metabolism. To ensure that these findings are accurate and applicable to a wider population, it is essential to conduct large-scale studies involving multiple centers. Combining genomic data with clinical outcomes and long-term surveillance studies will provide a more comprehensive understanding of how SNPs affect imatinib response in specific populations. Moreover, advancements in pharmacogenomics may lead to the development of predictive models that can guide personalized treatment plans for CML.
There is a potential way to improve the outcomes of CML treatment by considering the relationship between SNPs and low imatinib levels in patients. Despite these challenges, continuous research and collaboration among healthcare experts can help to develop personalized treatment plans. This, in turn, can lead to better safety and effectiveness of Imatinib treatment for patients suffering from chronic myeloid leukaemia.
The study protocol was approved by the Islamic International Medical Collegeethical review committee Ref#Riphah/IIMC/ERC/19/0265 dated 25th July 2019. The study protocol were in accordance with the declaration of Helsinki and guidelines provided by good clinical practice.
Informed written consent was obtained from all participants before the start of the study.
Fig share: Data of demographic features of Chronic myeloid leukaemia patients taking Imatinib (400 mg) with genetic and allelic frequencies of rs2242480 is available with: https://doi.org/10.6084/m9.figshare.25448260.v1 (Khan, 2024a).
Fig share: Data for Genetic models for rs2242280 in CML patients taking Imatinib (400 mg) is available with https://doi.org/10.6084/m9.figshare.25458964.v1 (Khan, 2024b).
Fig share: Figure for Plasma trough concentration of Imatinib (400 mg) in genotypes of rs2242480 is available with https://doi.org/10.6084/m9.figshare.25458973.v1 (Khan, 2024c).
Fig share: SPSS Data for rs2242480 Genotype is shared with link https://doi.org/10.6084/m9.figshare.25464859.v1 (Khan, 2024d).
Data are available under the terms of the Creative Commons Attribution: licence CC0.
Fig share: Strobe guidelines: Impact of CYP3A4 rs2242480 polymorphisms on Imatinib HPLC determined trough concentration and response in chronic myeloid leukaemia in Pakistani patients: DOI https://doi.org/10.6084/m9. figshare.25465027.v1 (Khan, 2024e).
Data are available under the terms of the Creative Commons Attribution: licence CC0
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Pediatric oncology and hematology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 22 Apr 24 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)