Keywords
Pectobcterium brasiliense, Histidine Kinase, Genome, Plant pathogen, Potato disease
This article is included in the Genomics and Genetics gateway.
A compilation of kinase genes of Pectobacterium brasilense is presented. The genes were mined from a complete genome sequence of strain BD163 isolated from potato in Tshwane, South Africa. This work is an effort to collate knowledge on P. brasiliense which has become a worldwide pathogen of potatoes and other vegetables such as tomato. Histidine kinases of P. brasiliense can be targets of bacterial control agents to limit the growth, spread and pathogenicity of P. brasiliense.
The whole genome sequence of strain BD163 of P. brasiliense deposited on www.ncbi.nlm.nih.gov was downloaded, and individual kinase genes were searched and listed with their properties, namely, locus tag, protein accession number and annotation.
This is the only compilation of histidine kinase genes for P. brasiliense.
Pectobcterium brasiliense, Histidine Kinase, Genome, Plant pathogen, Potato disease
Major Differences Between Current and Previous Versions:
In this revised version of our manuscript, we have incorporated several significant updates based on the valuable feedback from the reviewers. Firstly, we have expanded the introduction to include detailed information on using histidine kinase inhibitors as a potential control strategy for bacterial infections. This addition highlights the role of histidine kinase inhibitors in altering the expression of virulence factors and the bacteria's ability to adapt to the host environment, thereby offering a promising strategy to control bacterial infections. Relevant references have been added to support this concept, including works by Bem et al. (2015) and Espinasse et al. (2023).
Another significant update is the revision of the schematic representation of histidine kinase in Figure 1. We've made these changes to enhance clarity and aid in understanding the phosphate transfer process. Specifically, we've replaced the letter "A" with "D" in the legend to maintain consistency, and we've clarified the description to accurately reflect the phosphate transfer process from the histidine kinase to the response regulator (RR).
Additionally, we have updated the table title to summarise histidine kinase genes. The new title, "Putative Genes Related to Histidine Kinase Characteristics in the Genome of Pectobacterium brasiliense Strain BD163", more accurately describes the content of the table. This revision ensures that the reader can understand the table's content at a glance.
These revisions aim to enhance the manuscript's overall clarity, depth, and scientific accuracy by addressing the insightful comments and suggestions provided by the reviewers.
See the authors' detailed response to the review by Iris Yedidia
Histidine kinases are enzymes that catalyse phosphorylation reactions involving histidine (Figure 1) (Catlett et al., 2003). These kinases are essential for signal transduction and bacterial adaptability to new environments, and they are and widely distributed in bacteria. The function performed by histidine kinases is critical to the function of an organism, and therefore targeting histidine kinases in Pectobacterium brasiliense to limit its growth, spread, and pathogenicity may help reduce its impact as a plant pathogen. Pectobcterium brasiliense is a crops-tissue-macerating pathogen of different crops, and it has gained prominence with recent reports from multiple hosts in Taiwan, China and Korea (Li et al., 2023; Park et al., 2022; Song et al., 2023). Similar to other bacteria, P. brasilense gains entry into the plant through natural openings or wounds. It spreads throughout the intercellular spaces, and upon close contact with target plant cells, it releases a barrage of tissue macerating effectors to disrupt cells and after that survives on the oozing sap. The outside appearance of affected tissues is discoloured, and as the disease worsens, the plant weakens and eventually succumbs to infection. If not interrupted by disease control agents, P. brasiliense may cause unprecedented damage to individual plants and the plants close to the initially infected plant. With the discouraged use of antibiotics ways to control P. brasiliense are limited and therefore new drug targets are necessary. Histidine kinases inhibitors are widely employed as agents in drug design and development. Inhibition of bacterial virulence and pathogenicity was the demonstrated previously using HKs inhibitors (Bem et al., 2015) by changing the expression of virulence factors and the ability of bacteria to adapt to the host environment (Espinasse et al., 2023). Before efforts to design kinase targets in P. brasiliense, it is necessary to first compile all histidine kinases in P. brasiliense. This present work aimed to list all histidine kinases present in P. brasiliense to enable future efforts for design, formulation and testing of histidine kinase inhibitors. We created a list of histidine kinase genes of P. brasilense, strain BD163, whose complete genome sequence is published by Rauwane et al. (2023).
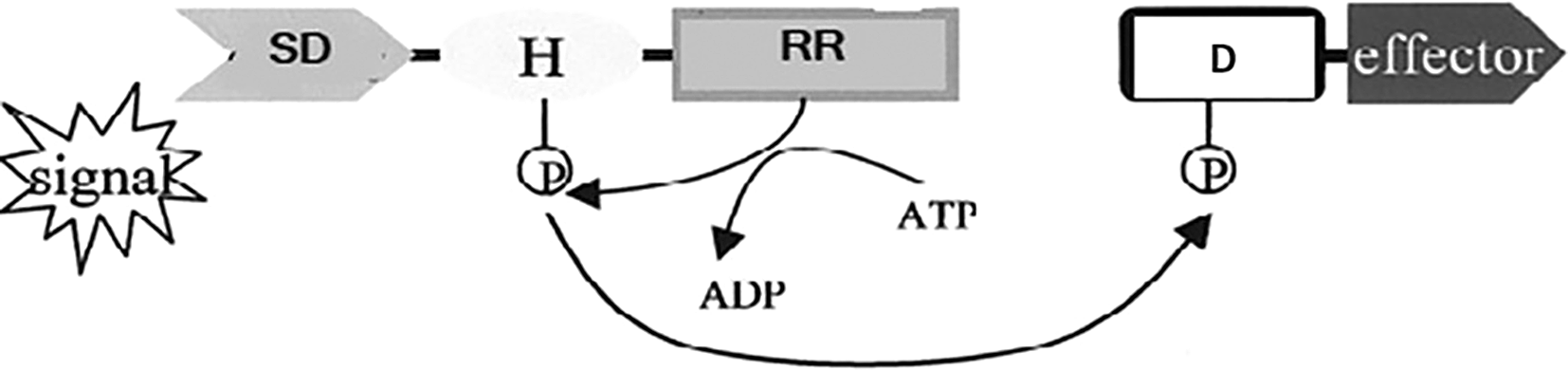
Phosphorylation of a histidine (H) residue is triggered by an external stimulus and detected by the sensor domain. The effector domain is activated when phosphate is transferred to a conserved aspartic acid residue (D) in the RR receiver domain. This leads to an action such as transcriptional activation or a MAP kinase (MAPK) cascade.
The genome sequence of strain BD163 of P. brasiliense, BioProject no. PRJNA798891, with raw reads under accession no. JAKNTB000000000, BioSample number SAMN25132833, and GenBank assembly accession number GCA_022172285.1 were downloaded, and individual histidine kinase genes were searched using the standard search function. To create the histidine kinase gene catalogue, the genes were listed with their characteristics: locus tag, protein accession number and annotation (Table 1).
To validate the histidine kinase gene repertoire of P. brasiliense, a comparative analysis was conducted by comparing the identified histidine kinase genes from strain BD163 with other available Pectobacterium genomes in the NCBI database. This analysis aimed to explore the conservation and diversity of histidine kinase genes within the genus Pectobacterium. The histidine kinase genes of three Pectobacterium strains (Pectobacterium carotovorum NX18131, Pectobacterium atrosepticum PB72, and Pectobacterium parmentieri IFB5626) were compared as part of a genome comparison study. The histidine kinase genes were found following the procedures outlined in the methods section. Based on the comparison results, it was found that similar to BD163, strains of P. carotovorum NX18131 and P. atrosepticum PB72 each contained 24 histidine kinase genes, whereas strain IFB5626 of P. parmentieri had just 20. These values show histidine kinase gene frequency differences among Pectobacterium species.
The genome sequence of strain BD163 of P. brasiliense, BioProject no. PRJNA798891, with raw reads under accession no. JAKNTB000000000, BioSample number SAMN25132833, and GenBank assembly accession number GCA_022172285.1 is obtainable from www.ncbi.nih.nlm.gov.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Partly
References
1. Qin Z, Zhang J, Xu B, Chen L, et al.: Structure-based discovery of inhibitors of the YycG histidine kinase: new chemical leads to combat Staphylococcus epidermidis infections.BMC Microbiol. 2006; 6: 96 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Plant pathology, Plant microbe interaction, bacterial virulence,
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |
|---|---|
| 1 | |
|
Version 2 (revision) 17 Jun 24 |
read |
|
Version 1 23 Apr 24 |
read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)